Bridge-1, a novel PDZ-domain coactivator of E2A-mediated regulation of insulin gene transcription
- PMID: 10567574
- PMCID: PMC84960
- DOI: 10.1128/MCB.19.12.8492
Bridge-1, a novel PDZ-domain coactivator of E2A-mediated regulation of insulin gene transcription
Abstract
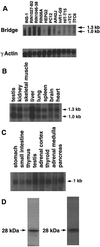
Proteins in the E2A family of basic helix-loop-helix transcription factors are important in a wide spectrum of physiologic processes as diverse as neurogenesis, myogenesis, lymphopoeisis, and sex determination. In the pancreatic beta cell, E2A proteins, in combination with tissue-specific transcription factors, regulate expression of the insulin gene and other genes critical for beta-cell function. By yeast two-hybrid screening of a cDNA library prepared from rat insulinoma (INS-1) cells, we identified a novel protein, Bridge-1, that interacts with E2A proteins and functions as a coactivator of gene transcription mediated by E12 and E47. Bridge-1 contains a PDZ-like domain, a domain known to be involved in protein-protein interactions. Bridge-1 is highly expressed in pancreatic islets and islet cell lines and the expression pattern is primarily nuclear. The interaction of Bridge-1 with E2A proteins is further demonstrated by coimmunoprecipitation of in vitro-translated Bridge-1 with E12 or E47 and by mammalian two-hybrid studies. The PDZ-like domain of Bridge-1 is required for interaction with the carboxy terminus of E12. In both yeast and mammalian two-hybrid interaction studies, Bridge-1 mutants lacking an intact PDZ-like domain interact poorly with E12. An E12 mutant (E12DeltaC) lacking the carboxy-terminal nine amino acids shows impaired interaction with Bridge-1. Bridge-1 has direct transactivational activity, since a Gal4 DNA-binding domain-Bridge-1 fusion protein transactivates a Gal4CAT reporter. Bridge-1 also functions as a coactivator by enhancing E12- or E47-mediated activation of a rat insulin I gene minienhancer promoter-reporter construct in transient-transfection experiments. Substitution of the mutant E12DeltaC for E12 reduces the coactivation of the rat insulin I minienhancer by Bridge-1. Inactivation of endogenous Bridge-1 in insulinoma (INS-1) cells by expression of a Bridge-1 antisense RNA diminishes rat insulin I promoter activity. Bridge-1, by utilizing its PDZ-like domain to interact with E12, may provide a new mechanism for the coactivation and regulation of transcription of the insulin gene.
Figures














References
-
- Anand G, Yin X, Shahidi A K, Grove L, Prochownik E V. Novel regulation of the helix-loop-helix protein Id1 by S5a, a subunit of the 26S proteasome. J Biol Chem. 1997;272:19140–19151. - PubMed
-
- Asfari M, Janjic D, Meda P, Li G, Halban P A, Wollheim C B. Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology. 1992;130:167–178. - PubMed
-
- Bain G, Maandag E C, Izon D J, Amsen D, Kruisbeek A M, Weintraub B C, Krop I, Schlisssel M S, Feeney A J, van Room M, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. - PubMed
-
- Cabral J H M, Petosa C, Sutcliffe M J, Raza S, Byron O, Poy F, Marfatia S M, Chishti A H, Liddington R C. Crystal structure of a PDZ domain. Nature. 1996;382:649–652. - PubMed
-
- Cho K-O, Hunt C A, Kennedy M B. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
