Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S phase
- PMID: 10567581
- PMCID: PMC84981
- DOI: 10.1128/MCB.19.12.8570
Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S phase
Abstract
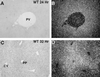
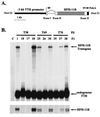
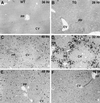
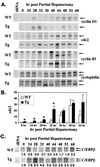
Two-thirds partial hepatectomy (PH) induces differentiated cells in the liver remnant to proliferate and regenerate to its original size. The proliferation-specific HNF-3/fork head homolog-11B protein (HFH-11B; also known as Trident and Win) is a family member of the winged helix/fork head transcription factors and in regenerating liver its expression is reactivated prior to hepatocyte entry into DNA replication (S phase). To examine whether HFH-11B regulates hepatocyte proliferation during liver regeneration, we used the -3-kb transthyretin (TTR) promoter to create transgenic mice that displayed ectopic hepatocyte expression of HFH-11B. Liver regeneration studies with the TTR-HFH-11B mice demonstrate that its premature expression resulted in an 8-h acceleration in the onset of hepatocyte DNA replication and mitosis. This liver regeneration phenotype is associated with protracted expression of cyclin D1 and C/EBPbeta, which are involved in stimulating DNA replication and premature expression of M phase promoting cyclin B1 and cdc2. Consistent with the early hepatocyte entry into S phase, regenerating transgenic livers exhibited earlier expression of DNA repair genes (XRCC1, mHR21spA, and mHR23B). Furthermore, in nonregenerating transgenic livers, ectopic HFH-11B expression did not elicit abnormal hepatocyte proliferation, a finding consistent with the retention of the HFH-11B transgene protein in the cytoplasm. We found that nuclear translocation of the HFH-11B transgene protein requires mitogenic signalling induced by PH and that its premature availability in regenerating transgenic liver allowed nuclear translocation to occur 8 h earlier than in wild type.
Figures




References
-
- Brenner D A. Signal transduction during liver regeneration. J Gastroenterol Hepatol. 1998;13:S93–S95. - PubMed
-
- Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. - PubMed
-
- Clark K L, Halay E D, Lai E, Burley S K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. - PubMed
-
- Corral M, Paris B, Guguen-Guillouzo C, Corcos D, Kruh J, Defer N. Increased expression of the N-myc gene during normal and neoplastic rat liver growth. Exp Cell Res. 1988;174:107–115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
