NF-Y associates with H3-H4 tetramers and octamers by multiple mechanisms
- PMID: 10567583
- PMCID: PMC84987
- DOI: 10.1128/MCB.19.12.8591
NF-Y associates with H3-H4 tetramers and octamers by multiple mechanisms
Abstract
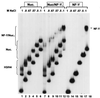
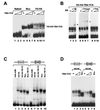
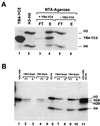
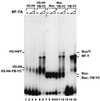
NF-Y is a CCAAT-binding trimer with two histonic subunits, NF-YB and NF-YC, resembling H2A-H2B. We previously showed that the short conserved domains of NF-Y efficiently bind to the major histocompatibility complex class II Ea Y box in DNA nucleosomized with purified chicken histones. Using wild-type NF-Y and recombinant histones, we find that NF-Y associates with H3-H4 early during nucleosome assembly, under conditions in which binding to naked DNA is not observed. In such assays, the NF-YB-NF-YC dimer forms complexes with H3-H4, for whose formation the CCAAT box is not required. We investigated whether they represent octamer-like structures, using DNase I, micrococcal nuclease, and exonuclease III, and found a highly positioned nucleosome on Ea, whose boundaries were mapped; addition of NF-YB-NF-YC does not lead to the formation of octameric structures, but changes in the digestion patterns are observed. NF-YA can bind to such preformed DNA complexes in a CCAAT-dependent way. In the absence of DNA, NF-YB-NF-YC subunits bind to H3-H4, but not to H2A-H2B, through the NF-YB histone fold. These results indicate that (i) the NF-Y histone fold dimer can efficiently associate DNA during nucleosome formation; (ii) it has an intrinsic affinity for H3-H4 but does not form octamers; and (iii) the interactions between NF-YA, NF-YB-NF-YC, and H3-H4 or nucleosomes are not mutually exclusive. Thus, NF-Y can intervene at different steps during nucleosome formation, and this scenario might be paradigmatic for other histone fold proteins involved in gene regulation.
Figures






References
-
- Alevizopoulos A, Dusserre Y, Tsai-Pflugfelder M, von der Weid T, Whali W, Mermod N. A proline-rich TGF-β-responsive transcriptional activator interacts with histones H3. Genes Dev. 1995;9:3051–3066. - PubMed
-
- Bellorini M, Zemzoumi K, Farina A, Berthelsen J, Piaggio G, Mantovani R. Cloning and expression of human NF-YC. Gene. 1997;193:119–125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
