Exploring expression data: identification and analysis of coexpressed genes
- PMID: 10568750
- PMCID: PMC310826
- DOI: 10.1101/gr.9.11.1106
Exploring expression data: identification and analysis of coexpressed genes
Abstract
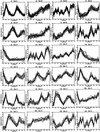
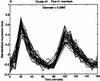
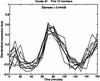
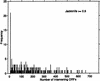
Analysis procedures are needed to extract useful information from the large amount of gene expression data that is becoming available. This work describes a set of analytical tools and their application to yeast cell cycle data. The components of our approach are (1) a similarity measure that reduces the number of false positives, (2) a new clustering algorithm designed specifically for grouping gene expression patterns, and (3) an interactive graphical cluster analysis tool that allows user feedback and validation. We use the clusters generated by our algorithm to summarize genome-wide expression and to initiate supervised clustering of genes into biologically meaningful groups.
Figures





Similar articles
-
Clustering of gene expression data: performance and similarity analysis.BMC Bioinformatics. 2006 Dec 12;7 Suppl 4(Suppl 4):S19. doi: 10.1186/1471-2105-7-S4-S19. BMC Bioinformatics. 2006. PMID: 17217511 Free PMC article.
-
Attribute clustering for grouping, selection, and classification of gene expression data.IEEE/ACM Trans Comput Biol Bioinform. 2005 Apr-Jun;2(2):83-101. doi: 10.1109/TCBB.2005.17. IEEE/ACM Trans Comput Biol Bioinform. 2005. PMID: 17044174
-
Finding dominant sets in microarray data.Front Biosci. 2005 Sep 1;10:3068-77. doi: 10.2741/1763. Front Biosci. 2005. PMID: 15970561
-
Overview on techniques in cluster analysis.Methods Mol Biol. 2010;593:81-107. doi: 10.1007/978-1-60327-194-3_5. Methods Mol Biol. 2010. PMID: 19957146 Review.
-
Comparing algorithms for clustering of expression data: how to assess gene clusters.Methods Mol Biol. 2009;541:479-509. doi: 10.1007/978-1-59745-243-4_21. Methods Mol Biol. 2009. PMID: 19381534 Review.
Cited by
-
Effects of 16S rDNA sampling on estimates of the number of endosymbiont lineages in sucking lice.PeerJ. 2016 Jul 19;4:e2187. doi: 10.7717/peerj.2187. eCollection 2016. PeerJ. 2016. PMID: 27547523 Free PMC article.
-
Alternative somatic and germline gene-regulatory strategies during starvation-induced developmental arrest.Cell Rep. 2022 Oct 11;41(2):111473. doi: 10.1016/j.celrep.2022.111473. Cell Rep. 2022. PMID: 36223742 Free PMC article.
-
A new method for alignment of LC-MALDI-TOF data.Proteome Sci. 2011 Oct 14;9 Suppl 1(Suppl 1):S10. doi: 10.1186/1477-5956-9-S1-S10. Proteome Sci. 2011. PMID: 22166061 Free PMC article.
-
Mass spectrometry imaging of lipids: untargeted consensus spectra reveal spatial distributions in Niemann-Pick disease type C1.J Lipid Res. 2018 Dec;59(12):2446-2455. doi: 10.1194/jlr.D086090. Epub 2018 Sep 28. J Lipid Res. 2018. PMID: 30266834 Free PMC article.
-
Microarray analysis: a novel research tool for cardiovascular scientists and physicians.Heart. 2003 Jun;89(6):597-604. doi: 10.1136/heart.89.6.597. Heart. 2003. PMID: 12748210 Free PMC article. Review.
References
-
- Cho R, Campbell M, Winzeler E, Steinmetz L, Conway A, Wodicka L, Wolfsberg T, Gabrielian A, Landsman D, Lockhart D, Davis R. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. - PubMed
-
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. - PubMed
-
- DeRisi J, Iyer V, Brown P. Exploring the metabolic and genetic control of gene expression on a genome scale. Science. 1997;278:680–686. - PubMed
-
- Efron B. The Jackknife, the Bootstrap, and Other Resampling Plans. CBMS-NSF Regional Conference Series in Applied Mathematics; 38. Society for Industrial & Applied Mathematics; 1982.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
