Detecting and analyzing DNA sequencing errors: toward a higher quality of the Bacillus subtilis genome sequence
- PMID: 10568751
- PMCID: PMC310837
- DOI: 10.1101/gr.9.11.1116
Detecting and analyzing DNA sequencing errors: toward a higher quality of the Bacillus subtilis genome sequence
Abstract
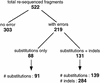
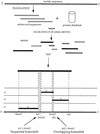
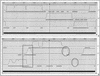
During the determination of a DNA sequence, the introduction of artifactual frameshifts and/or in-frame stop codons in putative genes can lead to misprediction of gene products. Detection of such errors with a method based on protein similarity matching is only possible when related sequences are available in databases. Here, we present a method to detect frameshift errors in DNA sequences that is based on the intrinsic properties of the coding sequences. It combines the results of two analyses, the search for translational initiation/termination sites and the prediction of coding regions. This method was used to screen the complete Bacillus subtilis genome sequence and the regions flanking putative errors were resequenced for verification. This procedure allowed us to correct the sequence and to analyze in detail the nature of the errors. Interestingly, in several cases in-frame termination codons or frameshifts were not sequencing errors but confirmed to be present in the chromosome, indicating that the genes are either nonfunctional (pseudogenes) or subject to regulatory processes such as programmed translational frameshifts. The method can be used for checking the quality of the sequences produced by any prokaryotic genome sequencing project.
Figures




References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Atkins JF, Weiss RB, Thompson S, Gesteland RF. Towards a genetic dissection of the basis of triplet decoding, and its natural subversion: Programmed reading frame shift and hops. Annu Rev Genet. 1991;25:201–228. - PubMed
-
- Atkins JF, Böck A, Matsufuji S, Gesteland RF. Dynamics of the genetic code. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA world. 2nd edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 637–673.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
