Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes
- PMID: 10570128
- PMCID: PMC24120
- DOI: 10.1073/pnas.96.24.13656
Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes
Abstract
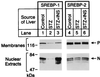
Sterol regulatory element binding proteins (SREBPs) enhance transcription of genes encoding enzymes of cholesterol and fatty acid biosynthesis and uptake. In the current experiments, we observed a decline in the mRNA encoding one SREBP isoform, SREBP-1c, in the livers of rats that were rendered diabetic by treatment with streptozotocin. There was no change in the mRNA encoding SREBP-1a, which is derived from the same gene as SREBP-1c but uses a different promoter. The ratio of SREBP-1c:1a transcripts fell 25-fold from 5:1 in control rats to 0.2:1 in the diabetic animals. The SREBP-1c mRNA rose nearly to normal, and the 1c:1a ratio increased 17-fold when the diabetic rats were treated for 6 h with insulin. These treatments produced no change in the mRNA for SREBP-2, which is encoded by a separate gene. The SREBP-1c mRNA also fell selectively in freshly isolated rat hepatocytes and rose when the cells were treated with insulin. Considered together with recent data on hepatocytes [Foretz, M., Pacot, C., Dugal, I., et al. (1999) Mol. Cell. Biol. 19, 3760-3768], the current in vivo studies suggest that insulin may stimulate lipid synthesis in the liver by selectively inducing transcription of the SREBP-1c gene.
Figures



Comment in
-
ADD-1 provides major new insight into the mechanism of insulin action.Proc Natl Acad Sci U S A. 1999 Dec 7;96(25):14191-2. doi: 10.1073/pnas.96.25.14191. Proc Natl Acad Sci U S A. 1999. PMID: 10588675 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

