Recurrent invasion and extinction of a selfish gene
- PMID: 10570167
- PMCID: PMC24159
- DOI: 10.1073/pnas.96.24.13880
Recurrent invasion and extinction of a selfish gene
Abstract
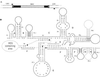
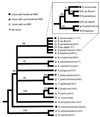
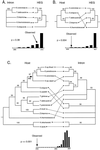
Homing endonuclease genes show super-Mendelian inheritance, which allows them to spread in populations even when they are of no benefit to the host organism. To test the idea that regular horizontal transmission is necessary for the long-term persistence of these genes, we surveyed 20 species of yeasts for the omega-homing endonuclease gene and associated group I intron. The status of omega could be categorized into three states (functional, nonfunctional, or absent), and status was not clustered on the host phylogeny. Moreover, the phylogeny of omega differed significantly from that of the host, strong evidence of horizontal transmission. Further analyses indicate that horizontal transmission is more common than transposition, and that it occurs preferentially between closely related species. Parsimony analysis and coalescent theory suggest that there have been 15 horizontal transmission events in the ancestry of our yeast species, through simulations indicate that this value is probably an underestimate. Overall, the data support a cyclical model of invasion, degeneration, and loss, followed by reinvasion, and each of these transitions is estimated to occur about once every 2 million years. The data are thus consistent with the idea that frequent horizontal transmission is necessary for the long-term persistence of homing endonuclease genes, and further, that this requirement limits these genes to organisms with easily accessible germ lines. The data also show that mitochondrial DNA sequences are transferred intact between yeast species; if other genes do not show such high levels of horizontal transmission, it would be due to lack of selection, rather than lack of opportunity.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

