Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes
- PMID: 10570185
- PMCID: PMC24177
- DOI: 10.1073/pnas.96.24.13983
Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes
Abstract
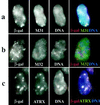
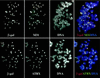
ATRX is a member of the SNF2 family of helicase/ATPases that is thought to regulate gene expression via an effect on chromatin structure and/or function. Mutations in the hATRX gene cause severe syndromal mental retardation associated with alpha-thalassemia. Using indirect immunofluorescence and confocal microscopy we have shown that ATRX protein is associated with pericentromeric heterochromatin during interphase and mitosis. By coimmunofluorescence, ATRX localizes with a mouse homologue of the Drosophila heterochromatic protein HP1 in vivo, consistent with a previous two-hybrid screen identifying this interaction. From the analysis of a trap assay for nuclear proteins, we have shown that the localization of ATRX to heterochromatin is encoded by its N-terminal region, which contains a conserved plant homeodomain-like finger and a coiled-coil domain. In addition to its association with heterochromatin, at metaphase ATRX clearly binds to the short arms of human acrocentric chromosomes, where the arrays of ribosomal DNA are located. The unexpected association of a putative transcriptional regulator with highly repetitive DNA provides a potential explanation for the variability in phenotype of patients with identical mutations in the ATRX gene.
Figures




References
-
- Gibbons R J, Brueton L, Buckle V J, Burn J, Clayton-Smith J, Davison B C C, Gardner R J M, Homfray T, Kearney L, Kingston H M, et al. Am J Med Genet. 1995;55:288–299. - PubMed
-
- Gibbons R J, Picketts D J, Villard L, Higgs D R. Cell. 1995;80:837–845. - PubMed
-
- Picketts D J, Higgs D R, Bachoo S, Blake D J, Quarrell O W J, Gibbons R J. Hum Mol Genet. 1996;5:1899–1907. - PubMed
-
- Kingston R E, Bunker C A, Imbalzano A N. Genes Dev. 1996;10:905–920. - PubMed
-
- Gibbons R J, Bachoo S, Picketts D J, Aftimos S, Asenbauer B, Bergoffen J, Berry S A, Dahl N, Fryer A, Keppler K, et al. Nat Genet. 1997;17:146–148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

