Differentially expressed protein Pdcd4 inhibits tumor promoter-induced neoplastic transformation
- PMID: 10570194
- PMCID: PMC24186
- DOI: 10.1073/pnas.96.24.14037
Differentially expressed protein Pdcd4 inhibits tumor promoter-induced neoplastic transformation
Abstract
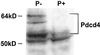
An mRNA differential display comparison of mouse JB6 promotion-sensitive (P+) and -resistant (P-) cells identified a novel gene product that inhibits neoplastic transformation. The JB6 P+ and P- cells are genetic variants that differ in their transformation response to tumor promoters; P+ cells form anchorage-independent colonies that are tumorigenic, and P- cells do not. A differentially displayed fragment, A7-1, was preferentially expressed in P- cells at levels >/=10-fold those in P+ cells, making its mRNA a candidate inhibitor of neoplastic transformation. An A7-1 cDNA was isolated that was identical to murine Pdcd4 gene cDNAs, also known as MA-3 or TIS, and analogous to human H731 and 197/15a. Until now, the function of the Pdcd4 protein has been unknown. Paralleling the mRNA levels, Pdcd4 protein levels were greater in P- than in P+ cells. Pdcd4 mRNA was also expressed at greater levels in the less progressed keratinocytes of another mouse skin neoplastic progression series. To test the hypothesis that Pdcd4 inhibits tumor promoter-induced transformation, stable cell lines expressing antisense Pdcd4 were generated from parental P- cells. The reduction of Pdcd4 proteins in antisense lines was accompanied by acquisition of a transformation-sensitive (P+) phenotype. The antisense-transfected cells were reverted to their initial P- phenotype by overexpression of a Pdcd4 sense fragment. These observations demonstrate that the Pdcd4 protein inhibits neoplastic transformation.
Figures



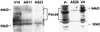


References
-
- Liang P, Pardee A B. Science. 1992;257:967–971. - PubMed
-
- Herschman H R. In: Advances in Regulation of Cell Growth, Volume 2; Cell Activation: Genetic Approaches. Mond J J, Cambier J C, Weiss A, editors. New York: Raven; 1991. pp. 83–117.
-
- Cmarik J L, Colburn N H. In: Food Factors for Cancer Prevention. Ohigashi H, Osawa T, Terao J, Watanabe S, Yoshikawa T, editors. Tokyo: Springer-Verlag; 1997. pp. 67–76.
-
- Colburn N H, Former B F, Nelson K A, Yuspa S H. Nature (London) 1979;281:589–591. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

