A periplasmic location is essential for the role of the ApbE lipoprotein in thiamine synthesis in Salmonella typhimurium
- PMID: 10572132
- PMCID: PMC103691
- DOI: 10.1128/JB.181.23.7285-7290.1999
A periplasmic location is essential for the role of the ApbE lipoprotein in thiamine synthesis in Salmonella typhimurium
Abstract
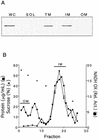
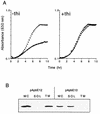
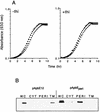
ApbE is a lipoprotein in Salmonella typhimurium, and mutants unable to make this protein have a reduced ability to make thiamine (vitamin B(1)) and require it as a supplement for optimal growth in minimal glucose medium. Polyclonal antibodies specific to ApbE were used to determine that wild-type ApbE is located exclusively in the inner membrane. The periplasmic, monotopic topology of ApbE was determined by using computer-based hydrophobicity plots, LacZ and PhoA gene fusions, and proteinase protection experiments. This extracellular location of ApbE is required for its function, since a cytoplasmic form (ApbE(cyto)) did not allow an apbE mutant to grow in the absence of thiamine. A periplasmic form of ApbE (ApbE(peri)) lacking the lipoprotein modification allowed an apbE mutant to grow in the absence of thiamine, indicating that soluble ApbE could function in thiamine synthesis and that lipoation and membrane association were not required. Alteration of the amino acid implicated in membrane sorting for other lipoproteins did not result in a relocalization of ApbE to the outer membrane, suggesting that additional sorting determinants exist for ApbE.
Figures

References
-
- Barik S. Site-directed mutagenesis by double polymerase chain reaction. Mol Biotechnol. 1995;3:1–7. - PubMed
-
- Bartolomé B, Jubete Y, Martinez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. - PubMed
-
- Begley T P, Downs D M, Ealick S, McLafferty F, van Loon D, Taylor S, Chiu H, Kinsland C, Reddick J, Xi J, Campobasso N. Thiamin synthesis in prokaryotes. Arch Microbiol. 1999;171:293–300. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

