A novel nitrate/nitrite permease in the marine Cyanobacterium synechococcus sp. strain PCC 7002
- PMID: 10572142
- PMCID: PMC103701
- DOI: 10.1128/JB.181.23.7363-7372.1999
A novel nitrate/nitrite permease in the marine Cyanobacterium synechococcus sp. strain PCC 7002
Abstract
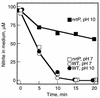
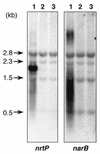
The nrtP and narB genes, encoding nitrate/nitrite permease and nitrate reductase, respectively, were isolated from the marine cyanobacterium Synechococcus sp. strain PCC 7002 and characterized. NrtP is a member of the major facilitator superfamily and is unrelated to the ATP-binding cassette-type nitrate transporters that previously have been described for freshwater strains of cyanobacteria. However, NrtP is similar to the NRT2-type nitrate transporters found in diverse organisms. An nrtP mutant strain consumes nitrate at a 4.5-fold-lower rate than the wild type, and this mutant grew exponentially on a medium containing 12 mM nitrate at a rate approximately 2-fold lower than that of the wild type. The nrtP mutant cells could not consume nitrite as rapidly as the wild type at pH 10, suggesting that NrtP also functions in nitrite uptake. A narB mutant was unable to grow on a medium containing nitrate as a nitrogen source, although this mutant could grow on media containing urea or nitrite with rates similar to those of the wild type. Exogenously added nitrite enhanced the in vivo activity of nitrite reductase in the narB mutant; this suggests that nitrite acts as a positive effector of nitrite reductase. Transcripts of the nrtP and narB genes were detected in cells grown on nitrate but were not detected in cells grown on urea or ammonia. Transcription of the nrtP and narB genes is probably controlled by the NtcA transcription factor for global nitrogen control. The discovery of a nitrate/nitrite permease in Synechococcus sp. strain PCC 7002 suggests that significant differences in nutrient transporters may occur in marine and freshwater cyanobacteria.
Figures

Similar articles
-
Nitrate/nitrite assimilation system of the marine picoplanktonic cyanobacterium Synechococcus sp. strain WH 8103: effect of nitrogen source and availability on gene expression.Appl Environ Microbiol. 2003 Dec;69(12):7009-18. doi: 10.1128/AEM.69.12.7009-7018.2003. Appl Environ Microbiol. 2003. PMID: 14660343 Free PMC article.
-
Role of NtcB in activation of nitrate assimilation genes in the cyanobacterium Synechocystis sp. strain PCC 6803.J Bacteriol. 2001 Oct;183(20):5840-7. doi: 10.1128/JB.183.20.5840-5847.2001. J Bacteriol. 2001. PMID: 11566981 Free PMC article.
-
Lack of control of nitrite assimilation by ammonium in an oceanic picocyanobacterium, Synechococcus sp. strain WH 8103.Appl Environ Microbiol. 2007 May;73(9):3028-33. doi: 10.1128/AEM.02606-06. Epub 2007 Mar 2. Appl Environ Microbiol. 2007. PMID: 17337543 Free PMC article.
-
Photosynthetic nitrate assimilation in cyanobacteria.Photosynth Res. 2005;83(2):117-33. doi: 10.1007/s11120-004-5830-9. Photosynth Res. 2005. PMID: 16143847 Review.
-
Nitrate assimilation by bacteria.Adv Microb Physiol. 1998;39:1-30, 379. doi: 10.1016/s0065-2911(08)60014-4. Adv Microb Physiol. 1998. PMID: 9328645 Review.
Cited by
-
A system-oriented strategy to enhance electron production of Synechocystis sp. PCC6803 in bio-photovoltaic devices: experimental and modeling insights.Sci Rep. 2021 Jun 10;11(1):12294. doi: 10.1038/s41598-021-91906-9. Sci Rep. 2021. PMID: 34112928 Free PMC article.
-
Contribution of protein synthesis depression to poly-β-hydroxybutyrate accumulation in Synechocystis sp. PCC 6803 under nutrient-starved conditions.Sci Rep. 2019 Dec 27;9(1):19944. doi: 10.1038/s41598-019-56520-w. Sci Rep. 2019. PMID: 31882765 Free PMC article.
-
Functional characterisation of substrate-binding proteins to address nutrient uptake in marine picocyanobacteria.Biochem Soc Trans. 2021 Dec 17;49(6):2465-2481. doi: 10.1042/BST20200244. Biochem Soc Trans. 2021. PMID: 34882230 Free PMC article. Review.
-
Computational inference and experimental validation of the nitrogen assimilation regulatory network in cyanobacterium Synechococcus sp. WH 8102.Nucleic Acids Res. 2006 Feb 10;34(3):1050-65. doi: 10.1093/nar/gkj496. Print 2006. Nucleic Acids Res. 2006. PMID: 16473855 Free PMC article.
-
Nitrate/nitrite assimilation system of the marine picoplanktonic cyanobacterium Synechococcus sp. strain WH 8103: effect of nitrogen source and availability on gene expression.Appl Environ Microbiol. 2003 Dec;69(12):7009-18. doi: 10.1128/AEM.69.12.7009-7018.2003. Appl Environ Microbiol. 2003. PMID: 14660343 Free PMC article.
References
-
- Bryant D A, Tandeau de Marsac N. Isolation of genes encoding components of photosynthetic apparatus. Methods Enzymol. 1988;167:755–765.
-
- Crawford N M, Glass A D M. Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 1998;3:389–395.
-
- Elhai J, Wolk C P. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allow cloning into long polylinkers. Gene. 1988;68:119–138. - PubMed
-
- Flores E, Guerrero M G, Losada M. Short-term ammonium inhibition of nitrate utilization by Anacystis nidulans and other cyanobacteria. Arch Microbiol. 1980;128:137–144.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

