Fetal Alz-50 clone 1, a novel zinc finger protein, binds a specific DNA sequence and acts as a transcriptional regulator
- PMID: 10575013
- PMCID: PMC3670955
- DOI: 10.1074/jbc.274.49.35262
Fetal Alz-50 clone 1, a novel zinc finger protein, binds a specific DNA sequence and acts as a transcriptional regulator
Abstract
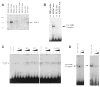
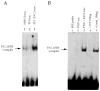
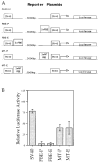
Fetal Alz-50 clone 1 (FAC1) is a novel, developmentally regulated gene that exhibits changes in protein expression and subcellular localization during neuronal development and neurodegeneration. To understand the functional implications of altered subcellular localization, we have established a normal cellular function of FAC1. The FAC1 amino acid sequence contains regional homology to transcriptional regulators. Using the polymerase chain reaction-assisted binding site selection assay, we have identified a DNA sequence recognized by recombinant FAC1. Mutation of any 2 adjacent base pairs in the identified binding site dramatically reduced the binding preference of FAC1, demonstrating that the binding is specific for the identified site. Nuclear extracts from neural and non-neural cell lines contained a DNA-binding activity with similar specificity and nucleotide requirements as the recombinant FAC1 protein. This DNA-binding activity can be attributed to FAC1 since it is dependent upon the presence of FAC1 and behaves identically on a nondenaturing polyacrylamide gel as transiently transfected FAC1. In NIH3T3 cells, luciferase reporter plasmids containing the identified binding site (CACAACAC) were repressed by cotransfected FAC1 whether the binding site was proximal or distal to the transcription initiation site. This study indicates that FAC1 is a DNA-binding protein that functions as a transcription factor when localized to the nucleus.
Figures



Similar articles
-
Fetal Alz-50 clone 1 interacts with the human orthologue of the Kelch-like Ech-associated protein.Biochemistry. 2004 Sep 28;43(38):12113-22. doi: 10.1021/bi0494166. Biochemistry. 2004. PMID: 15379550 Free PMC article.
-
Fetal Alz-50 clone 1 (FAC1) protein interacts with the Myc-associated zinc finger protein (ZF87/MAZ) and alters its transcriptional activity.Biochemistry. 2000 Mar 28;39(12):3206-15. doi: 10.1021/bi992211q. Biochemistry. 2000. PMID: 10727212 Free PMC article.
-
DNA binding activity of the fetal Alz-50 clone 1 (FAC1) protein is enhanced by phosphorylation.Biochem Biophys Res Commun. 1999 Jul 14;260(3):785-9. doi: 10.1006/bbrc.1999.0986. Biochem Biophys Res Commun. 1999. PMID: 10403843
-
FAC1, a novel gene identified with the monoclonal antibody Alz50, is developmentally regulated in human brain.Dev Neurosci. 1995;17(1):20-37. doi: 10.1159/000111270. Dev Neurosci. 1995. PMID: 7621746
-
Expression of the fetal Alz-50 clone 1 protein induces apoptotic cell death.Biochem Biophys Res Commun. 2005 Oct 21;336(2):490-5. doi: 10.1016/j.bbrc.2005.08.127. Biochem Biophys Res Commun. 2005. PMID: 16137655
Cited by
-
Opportunity knocks for uncovering the new function of an understudied nucleosome remodeling complex member, the bromodomain PHD finger transcription factor, BPTF.Curr Opin Chem Biol. 2021 Aug;63:57-67. doi: 10.1016/j.cbpa.2021.02.003. Epub 2021 Mar 8. Curr Opin Chem Biol. 2021. PMID: 33706239 Free PMC article. Review.
-
Functional repression of cAMP response element in 6-hydroxydopamine-treated neuronal cells.J Biol Chem. 2006 Jun 30;281(26):17870-81. doi: 10.1074/jbc.M602632200. Epub 2006 Apr 18. J Biol Chem. 2006. PMID: 16621793 Free PMC article.
-
mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival.Genes Dev. 2005 Jul 1;19(13):1581-95. doi: 10.1101/gad.1286905. Genes Dev. 2005. PMID: 15998811 Free PMC article.
-
The p53 homologue p73 accumulates in the nucleus and localizes to neurites and neurofibrillary tangles in Alzheimer disease brain.Neuropathol Appl Neurobiol. 2004 Feb;30(1):19-29. doi: 10.1046/j.0305-1846.2003.00496.x. Neuropathol Appl Neurobiol. 2004. PMID: 14720173 Free PMC article.
-
Fetal Alz-50 clone 1 interacts with the human orthologue of the Kelch-like Ech-associated protein.Biochemistry. 2004 Sep 28;43(38):12113-22. doi: 10.1021/bi0494166. Biochemistry. 2004. PMID: 15379550 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

