Localization of a gene for Duane retraction syndrome to chromosome 2q31
- PMID: 10577917
- PMCID: PMC1288374
- DOI: 10.1086/302656
Localization of a gene for Duane retraction syndrome to chromosome 2q31
Abstract
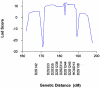
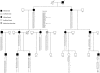
Duane retraction syndrome (DRS) is a congenital eye-movement disorder characterized by a failure of cranial nerve VI (the abducens nerve) to develop normally, resulting in restriction or absence of abduction, restricted adduction, and narrowing of the palpebral fissure and retraction of the globe on attempted adduction. DRS has a prevalence of approximately 0.1% in the general population and accounts for 5% of all strabismus cases. Undiagnosed DRS in children can lead to amblyopia, a permanent uncorrectable loss of vision. A large family with autosomal dominant DRS was examined and tested for genetic linkage. After exclusion of candidate regions previously associated with DRS, a genomewide search with highly polymorphic microsatellite markers was performed, and significant evidence for linkage was obtained at chromosome 2q31 (D2S2314 maximum LOD score 11.73 at maximum recombination fraction. 0). Haplotype analysis places the affected gene in a 17.8-cM region between the markers D2S2330 and D2S364. No recombinants were seen with markers between these two loci. The linked region contains the homeobox D gene cluster. Three of the genes within this cluster, known to participate in hindbrain development, were sequenced in affected and control individuals. Coding sequences for these genes were normal or had genetic alterations unlikely to be responsible for the DRS phenotype. Identifying the gene responsible for DRS may lead to an improved understanding of early cranial-nerve development.
Figures



References
Electronic-Database Information
-
- GenBank, http://www.ncbi.nlm.nih.gov/GenBank (for gene locations)
-
- Genetic Location Database, http://cedar.genetics.soton.ac.uk/ (for markers used for haplotype analysis)
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim (for Duane retraction syndrome [MIM 126800])
-
- PE Biosystems, http://www.pebio.com/ab/apply/dr/dra1a3.html (for markers used for linkage analysis), http://www2.perkin-elmer.com/ab/apply/dr/lmsv2/chrom2.html (for ABI PRISM mapping)
References
-
- Bedford M, Arman E, Orr-Urtreger A, Lonai P (1995) Analysis of the Hoxd-3 gene: structure and localization of its sense and natural antisense transcripts. DNA Cell Biol 14(4): 295–304 - PubMed
-
- Calabrese GSL, Morizio E, Franchi PG, Pompetti F, Mingarelli R, Marsilio T, Rocchi M, et al (1998) Detection of an insertion deletion of region 8q13-q21.2 in a patient with Duane syndrome: implications for mapping and cloning a Duane gene. Eur J Hum Genet 6(3): 187–193 - PubMed
-
- Carpenter EM, Goddard JM, Chisaka O, Manley NR, Capecchi MR (1993) Loss of Hox-A1 (Hox-1.6) function results in the reorganization of the murine hindbrain. Development 118:1063–1075 - PubMed
-
- Chew CK, Foster P, Hurst JA, Salmon JF (1995) Duane's retraction syndrome associated with chromosome 4q27-31 segment deletion. Am J Ophthalmol 119(6): 807–809 - PubMed
-
- Chung M, Stout JT, Borchert MS. Clinical diversity of hereditary duane's retraction syndrome. Ophthalmology (in press) - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

