The TOM core complex: the general protein import pore of the outer membrane of mitochondria
- PMID: 10579717
- PMCID: PMC2169338
- DOI: 10.1083/jcb.147.5.959
The TOM core complex: the general protein import pore of the outer membrane of mitochondria
Abstract
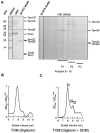
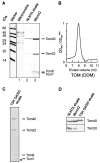
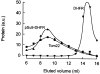
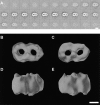
Translocation of nuclear-encoded preproteins across the outer membrane of mitochondria is mediated by the multicomponent transmembrane TOM complex. We have isolated the TOM core complex of Neurospora crassa by removing the receptors Tom70 and Tom20 from the isolated TOM holo complex by treatment with the detergent dodecyl maltoside. It consists of Tom40, Tom22, and the small Tom components, Tom6 and Tom7. This core complex was also purified directly from mitochondria after solubilization with dodecyl maltoside. The TOM core complex has the characteristics of the general insertion pore; it contains high-conductance channels and binds preprotein in a targeting sequence-dependent manner. It forms a double ring structure that, in contrast to the holo complex, lacks the third density seen in the latter particles. Three-dimensional reconstruction by electron tomography exhibits two open pores traversing the complex with a diameter of approximately 2.1 nm and a height of approximately 7 nm. Tom40 is the key structural element of the TOM core complex.
Figures




References
-
- Bains G., Lithgow T. The Tom channel in the outer membranealive and kicking. BioEssays. 1999;21:1–4. - PubMed
-
- Benz R., Janko K., Boos W., Läuger P. Formation of large, ion-permeable membrane channels by the matrix protein (porin) of Escherichia coli . Biochim. Biophys. Acta. 1978;511:305–319. - PubMed
-
- Brix J., Dietmeier K., Pfanner N. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J. Biol. Chem. 1997;272:20730–20735. - PubMed
-
- Cao W., Douglas M.G. Biogenesis of ISP6, a small carboxyl-terminal anchored protein of the receptor complex of the mitochondrial outer membrane. J. Biol. Chem. 1995;270:5674–5679. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

