Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig
- PMID: 10583991
- PMCID: PMC91731
- DOI: 10.1128/AEM.65.12.5372-5377.1999
Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig
Abstract
Random clones of 16S ribosomal DNA gene sequences were isolated after PCR amplification with eubacterial primers from total genomic DNA recovered from samples of the colonic lumen, colonic wall, and cecal lumen from a pig. Sequences were also obtained for cultures isolated anaerobically from the same colonic-wall sample. Phylogenetic analysis showed that many sequences were related to those of Lactobacillus or Streptococcus spp. or fell into clusters IX, XIVa, and XI of gram-positive bacteria. In addition, 59% of randomly cloned sequences showed less than 95% similarity to database entries or sequences from cultivated organisms. Cultivation bias is also suggested by the fact that the majority of isolates (54%) recovered from the colon wall by culturing were related to Lactobacillus and Streptococcus, whereas this group accounted for only one-third of the sequence variation for the same sample from random cloning. The remaining cultured isolates were mainly Selenomonas related. A higher proportion of Lactobacillus reuteri-related sequences than of Lactobacillus acidophilus- and Lactobacillus amylovorus-related sequences were present in the colonic-wall sample. Since the majority of bacterial ribosomal sequences recovered from the colon wall are less than 95% related to known organisms, the roles of many of the predominant wall-associated bacteria remain to be defined.
Figures
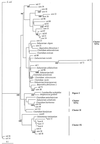
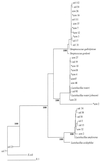

References
-
- Avgustin G, Wright F, Flint H J. Genetic diversity and phylogenetic relationships among strains of Prevotella (Bacteroides) ruminicola from the rumen. Int J Syst Bacteriol. 1994;47:284–288. - PubMed
-
- Bengmark S. Econutrition and health maintenance—a new concept to prevent GI inflammation, ulceration and sepsis. Clin Nutr. 1996;15:1–10. - PubMed
-
- Bryant M P. Commentary on the Hungate technique for cultivation of anaerobic bacteria. Am J Clin Nutr. 1972;25:1324–1328. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

