Quantification of bias related to the extraction of DNA directly from soils
- PMID: 10583997
- PMCID: PMC91737
- DOI: 10.1128/AEM.65.12.5409-5420.1999
Quantification of bias related to the extraction of DNA directly from soils
Abstract
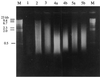
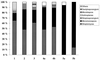
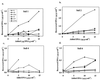
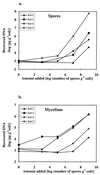
In recent years, several protocols based on the extraction of nucleic acids directly from the soil matrix after lysis treatment have been developed for the detection of microorganisms in soil. Extraction efficiency has often been evaluated based on the recovery of a specific gene sequence from an organism inoculated into the soil. The aim of the present investigation was to improve the extraction, purification, and quantification of DNA derived from as large a portion of the soil microbial community as possible, with special emphasis placed on obtaining DNA from gram-positive bacteria, which form structures that are difficult to disrupt. Furthermore, we wanted to identify and minimize the biases related to each step in the procedure. Six soils, covering a range of pHs, clay contents, and organic matter contents, were studied. Lysis was carried out by soil grinding, sonication, thermal shocks, and chemical treatments. DNA was extracted from the indigenous microflora as well as from inoculated bacterial cells, spores, and hyphae, and the quality and quantity of the DNA were determined by gel electrophoresis and dot blot hybridization. Lysis efficiency was also estimated by microscopy and viable cell counts. Grinding increased the extracellular DNA yield compared with the yield obtained without any lysis treatment, but none of the subsequent treatments clearly increased the DNA yield. Phage lambda DNA was inoculated into the soils to mimic the fate of extracellular DNA. No more than 6% of this DNA could be recovered from the different soils. The clay content strongly influenced the recovery of DNA. The adsorption of DNA to clay particles decreased when the soil was pretreated with RNA in order to saturate the adsorption sites. We also investigated different purification techniques and optimized the PCR methods in order to develop a protocol based on hybridization of the PCR products and quantification by phosphorimaging.
Figures


References
-
- Clegg C D, Ritz K, Griffiths B S. Direct extraction of microbial community DNA from humified upland soils. Lett Appl Microbiol. 1997;25:30–33. - PubMed
-
- Clerc-Bardin, S., J.-L. Pernodet, Å. Frostegård, and P. Simonet. Development of a conditional suicide system for a Streptomyces lividans strain and its use to investigate conjugative transfer in soil. Submitted for publication.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

