Precise detection and tracing of Trichoderma hamatum 382 in compost-amended potting mixes by using molecular markers
- PMID: 10583998
- PMCID: PMC91738
- DOI: 10.1128/AEM.65.12.5421-5426.1999
Precise detection and tracing of Trichoderma hamatum 382 in compost-amended potting mixes by using molecular markers
Abstract
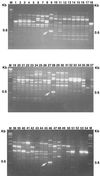
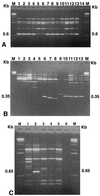
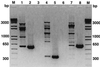
Randomly amplified polymorphic DNA (RAPD) analysis and the PCR assay were used in combination with dilution plating on a semiselective medium to detect and enumerate propagules of Trichoderma hamatum 382, a biocontrol agent utilized in compost-amended mixes. Distinct and reproducible fingerprints were obtained upon amplification of purified genomic DNA of T. hamatum 382 with the random primers OPE-16, OPH-19, and OPH-20. Three amplified DNA fragments of 0.35 (OPE-16(0.35)), 0.6 (OPH-19(0.6)), and 0.65 (OPH-20(0.65)) kb were diagnostic for T. hamatum 382, clearly distinguishing it from 53 isolates of four other Trichoderma spp. tested. Some isolates of T. hamatum shared these low-molecular-weight fragments with T. hamatum 382. However, RAPD analysis of isolates of T. hamatum with all three random primers used in consecutive PCR tests distinguished T. hamatum 382 from other isolates of T. hamatum. These three RAPD amplicons were cloned and sequenced, and pairs of oligonucleotide primers for each cloned fragment were designed. Use of the primers in the PCR assay resulted in the amplification of DNA fragments of the same size as the cloned RAPD fragments from genomic DNA of T. hamatum 382. A combination of dilution plating on a semiselective medium for Trichoderma spp. and PCR, with the RAPD primers OPH-19, OPE-16, and OPH-20 or the three sequence-characterized primers, was used successfully to verify the presence of T. hamatum 382 propagules in nine different soil, compost, and potting mix samples. All 23 Trichoderma isolates recovered on semiselective medium from commercial potting mixes fortified with T. hamatum 382 were identified as T. hamatum 382, whereas 274 Trichoderma isolates recovered from the other nine samples were negative in the PCR assay. Thus, this highly specific combination of techniques allowed detection and enumeration of propagules of T. hamatum 382 in fortified compost-amended potting mixes. Sequence-characterized amplified region markers also facilitated the development of a very simple procedure to amplify DNA of T. hamatum 382 directly from fortified compost-amended potting mixes.
Figures


References
-
- Arisan-Atac I, Heidenreich E, Kubicek C P. Randomly amplified polymorphic DNA fingerprinting identifies subgroups of Trichoderma viride and other Trichoderma spp. capable of chestnut blight biocontrol. FEMS Microbiol Lett. 1995;126:249–256. - PubMed
-
- Bissett J. A revision of the genus Trichoderma. III. Section Pachybasium. Can J Bot. 1991;69:2373–2417.
-
- Brisbane P G, Neate S M, Pankhurst C E, Scott N S, Thomas M R. Sequence-tagged site markers to identify Rhizoctonia solani AG4 or 8 infecting wheat in south Australia. Phytopathology. 1995;85:1423–1427.
-
- Chung Y R, Hoitink H A J. Interactions between thermophilic fungi and Trichoderma hamatum in suppression of Rhizoctonia damping-off in a bark compost-amended container medium. Phytopathology. 1990;80:73–77.
-
- Crowhurst R N, Hawthorne B T, Rikkerink E H A, Templeton M D. Differentiation of Fusarium solani f. sp. cucurbitae race 1 and 2 by random amplification of polymorphic DNA. Curr Genet. 1991;20:391–396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

