Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization
- PMID: 10584017
- PMCID: PMC91757
- DOI: 10.1128/AEM.65.12.5554-5563.1999
Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization
Abstract
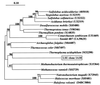
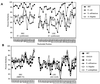
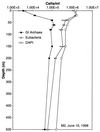
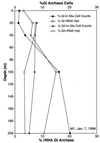
Fluorescent in situ hybridization (FISH) using rRNA-specific oligonucleotide probes has emerged as a popular technique for identifying individual microbial cells. In natural samples, however, the signal derived from fluor-labeled oligonucleotide probes often is undetectable above background fluorescence in many cells. To circumvent this difficulty, we applied fluorochrome-labeled polyribonucleotide probes to identify and enumerate marine planktonic archaea and bacteria. The approach greatly enhanced the sensitivity and applicability of FISH with seawater samples, allowing confident identification and enumeration of planktonic cells to ocean depths of 3,400 m. Quantitative whole-cell hybridization experiments using these probes accounted for 90 to 100% of the total 4',6-diamidino-2-phenylindole (DAPI)-stained cells in most samples. As predicted in a previous study (R. Massana, A. E. Murray, C. M. Preston, and E. F. DeLong, Appl. Environ. Microbiol. 63:50-56, 1997), group I and II marine archaea predominate in different zones in the water column, with maximal cell densities of 10(5)/ml. The high cell densities of archaea, extending from surface waters to abyssal depths, suggest that they represent a large and significant fraction of the total picoplankton biomass in coastal ocean waters. The data also show that the vast majority of planktonic prokaryotes contain significant numbers of ribosomes, rendering them easily detectable with polyribonucleotide probes. These results imply that the majority of planktonic cells visualized by DAPI do not represent lysed cells or "ghosts," as was suggested in a previous report.
Figures



References
-
- Amann R, Springer N, Ludwig W, Gortz H D, Schleifer K H. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991;351:161–164. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

