Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda
- PMID: 10585965
- PMCID: PMC98976
- DOI: 10.1128/MMBR.63.4.751-813.1999
Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda
Abstract
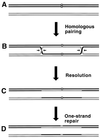
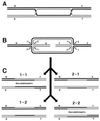
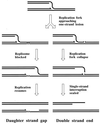
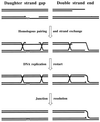
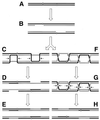
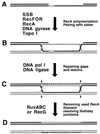
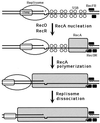
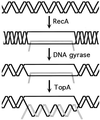
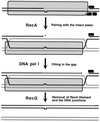
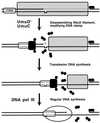
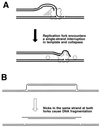
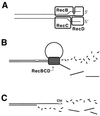
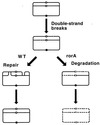
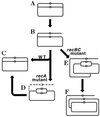
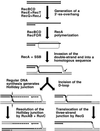
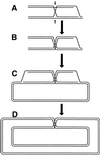
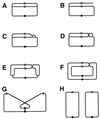
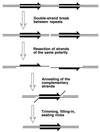
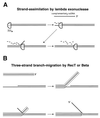
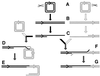
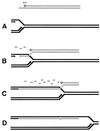
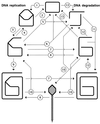
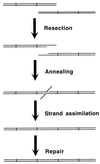
Although homologous recombination and DNA repair phenomena in bacteria were initially extensively studied without regard to any relationship between the two, it is now appreciated that DNA repair and homologous recombination are related through DNA replication. In Escherichia coli, two-strand DNA damage, generated mostly during replication on a template DNA containing one-strand damage, is repaired by recombination with a homologous intact duplex, usually the sister chromosome. The two major types of two-strand DNA lesions are channeled into two distinct pathways of recombinational repair: daughter-strand gaps are closed by the RecF pathway, while disintegrated replication forks are reestablished by the RecBCD pathway. The phage lambda recombination system is simpler in that its major reaction is to link two double-stranded DNA ends by using overlapping homologous sequences. The remarkable progress in understanding the mechanisms of recombinational repair in E. coli over the last decade is due to the in vitro characterization of the activities of individual recombination proteins. Putting our knowledge about recombinational repair in the broader context of DNA replication will guide future experimentation.
Figures

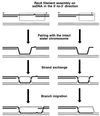
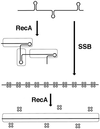
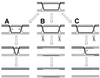
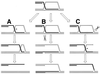
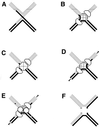
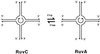
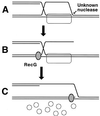

References
-
- Adzuma K. Stable synapsis of homologous DNA molecules mediated by the Escherichia coli RecA protein involves local exchange of DNA strands. Genes Dev. 1992;6:1679–1694. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

