Prions in Saccharomyces and Podospora spp.: protein-based inheritance
- PMID: 10585968
- PMCID: PMC98979
- DOI: 10.1128/MMBR.63.4.844-861.1999
Prions in Saccharomyces and Podospora spp.: protein-based inheritance
Abstract
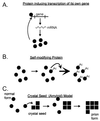
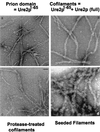
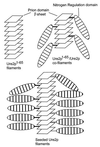
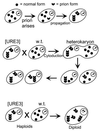
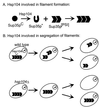
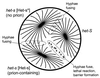
Genetic evidence showed two non-Mendelian genetic elements of Saccharomyces cerevisiae, called [URE3] and [PSI], to be prions of Ure2p and Sup35p, respectively. [URE3] makes cells derepressed for nitrogen catabolism, while [PSI] elevates the efficiency of weak suppressor tRNAs. The same approach led to identification of the non-Mendelian element [Het-s] of the filamentous fungus Podospora anserina, as a prion of the het-s protein. The prion form of the het-s protein is required for heterokaryon incompatibility, a normal fungal function, suggesting that other normal cellular functions may be controlled by prions. [URE3] and [PSI] involve a self-propagating aggregation of Ure2p and Sup35p, respectively. In vitro, Ure2p and Sup35p form amyloid, a filamentous protein structure, high in beta-sheet with a characteristic green birefringent staining by the dye Congo Red. Amyloid deposits are a cardinal feature of Alzheimer's disease, non-insulin-dependent diabetes mellitus, the transmissible spongiform encephalopathies, and many other diseases. The prion domain of Ure2p consists of Asn-rich residues 1 to 80, but two nonoverlapping fragments of the molecule can, when overproduced, induce the de nova appearance of [URE3]. The prion domain of Sup35 consists of residues 1 to 114, also rich in Asn and Gln residues. While runs of Asn and Gln are important for [URE3] and [PSI], no such structures are found in PrP or the Het-s protein. Either elevated or depressed levels of the chaperone Hsp104 interfere with propagation of [PSI]. Both [URE3] and [PSI] are cured by growth of cells in millimolar guanidine HCl. [URE3] is also cured by overexpression of fragments of Ure2p or fusion proteins including parts of Ure2p.
Figures




Similar articles
-
[URE3] and [PSI] are prions of yeast and evidence for new fungal prions.Curr Issues Mol Biol. 2000 Apr;2(2):51-9. Curr Issues Mol Biol. 2000. PMID: 11471564 Review.
-
[PSI] and [URE3] as yeast prions.Yeast. 1995 Dec;11(16):1671-85. doi: 10.1002/yea.320111609. Yeast. 1995. PMID: 8720070 Review.
-
Prions are affected by evolution at two levels.Cell Mol Life Sci. 2016 Mar;73(6):1131-44. doi: 10.1007/s00018-015-2109-6. Epub 2015 Dec 28. Cell Mol Life Sci. 2016. PMID: 26713322 Free PMC article. Review.
-
Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p.Mol Cell Biol. 2002 Jun;22(11):3590-8. doi: 10.1128/MCB.22.11.3590-3598.2002. Mol Cell Biol. 2002. PMID: 11997496 Free PMC article.
-
The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments.Proc Natl Acad Sci U S A. 1999 Feb 16;96(4):1498-503. doi: 10.1073/pnas.96.4.1498. Proc Natl Acad Sci U S A. 1999. PMID: 9990052 Free PMC article.
Cited by
-
[NSI (+)]: a novel non-Mendelian nonsense suppressor determinant in Saccharomyces cerevisiae.Curr Genet. 2010 Oct;56(5):467-78. doi: 10.1007/s00294-010-0314-2. Epub 2010 Jul 29. Curr Genet. 2010. PMID: 20668856
-
Nonself recognition is mediated by HET-C heterocomplex formation during vegetative incompatibility.EMBO J. 2002 Sep 16;21(18):4841-50. doi: 10.1093/emboj/cdf479. EMBO J. 2002. PMID: 12234924 Free PMC article.
-
Fungal prion HET-s as a model for structural complexity and self-propagation in prions.Proc Natl Acad Sci U S A. 2014 Apr 8;111(14):5201-6. doi: 10.1073/pnas.1322933111. Epub 2014 Mar 24. Proc Natl Acad Sci U S A. 2014. PMID: 24706820 Free PMC article.
-
Novel non-Mendelian determinant involved in the control of translation accuracy in Saccharomyces cerevisiae.Genetics. 2002 Jan;160(1):25-36. doi: 10.1093/genetics/160.1.25. Genetics. 2002. PMID: 11805042 Free PMC article.
-
Heterogeneous seeding of a prion structure by a generic amyloid form of the fungal prion-forming domain HET-s(218-289).J Biol Chem. 2013 Oct 11;288(41):29604-12. doi: 10.1074/jbc.M113.505511. Epub 2013 Aug 28. J Biol Chem. 2013. PMID: 23986444 Free PMC article.
References
-
- Aigle M, Lacroute F. Genetical aspects of [URE3], a non-Mendelian, cytoplasmically inherited mutation in yeast. Mol Gen Genet. 1975;136:327–335. - PubMed
-
- Alper T, Cramp W A, Haig D A, Clarke M C. Does the agent of scrapie replicate without nucleic acid? Nature. 1967;214:764–766. - PubMed
-
- Alper T, Haig D A, Clarke M C. The exceptionally small size of the scrapie agent. Biochem Biophys Res Commun. 1966;22:278–284. - PubMed
-
- Begueret J, Turq B, Clave C. Vegetative incompatibility in filamentous fungi: het genes begin to talk. Trends Genet. 1994;10:441–446. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

