Pex17p is required for import of both peroxisome membrane and lumenal proteins and interacts with Pex19p and the peroxisome targeting signal-receptor docking complex in Pichia pastoris
- PMID: 10588639
- PMCID: PMC25739
- DOI: 10.1091/mbc.10.12.4005
Pex17p is required for import of both peroxisome membrane and lumenal proteins and interacts with Pex19p and the peroxisome targeting signal-receptor docking complex in Pichia pastoris
Abstract
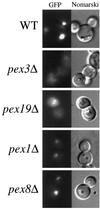
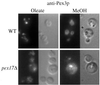
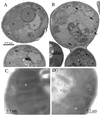
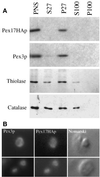
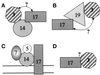
Pichia pastoris PEX17 was cloned by complementation of a peroxisome-deficient strain obtained from a novel screen for mutants disrupted in the localization of a peroxisomal membrane protein (PMP) reporter. PEX17 encodes a 267-amino-acid protein with low identity (18%) to the previously characterized Saccharomyces cerevisiae Pex17p. Like ScPex17p, PpPex17p contains a putative transmembrane domain near the amino terminus and two carboxyl-terminal coiled-coil regions. PpPex17p behaves as an integral PMP with a cytosolic carboxyl-terminal domain. pex17Delta mutants accumulate peroxisomal matrix proteins and certain integral PMPs in the cytosol, suggesting a critical role for Pex17p in their localization. Peroxisome remnants were observed in the pex17Delta mutant by morphological and biochemical means, suggesting that Pex17p is not absolutely required for remnant formation. Yeast two-hybrid analysis demonstrated that the carboxyl terminus of Pex19p was required for interaction with Pex17p lacking the carboxyl-terminal coiled-coil domains. Biochemical evidence confirmed the interaction between Pex19p and Pex17p. Additionally, Pex17p cross-linked to components of the peroxisome targeting signal-receptor docking complex, which unexpectedly contained Pex3p. Our evidence suggests the existence of distinct subcomplexes that contain separable pools of Pex3p, Pex19p, Pex17p, Pex14p, and the peroxisome targeting signal receptors. These distinct pools may serve different purposes for the import of matrix proteins or PMPs.
Figures






Similar articles
-
Pex19p interacts with Pex3p and Pex10p and is essential for peroxisome biogenesis in Pichia pastoris.Mol Biol Cell. 1999 Jun;10(6):1745-61. doi: 10.1091/mbc.10.6.1745. Mol Biol Cell. 1999. PMID: 10359594 Free PMC article.
-
Pex17p of Saccharomyces cerevisiae is a novel peroxin and component of the peroxisomal protein translocation machinery.J Cell Biol. 1998 Jan 12;140(1):49-60. doi: 10.1083/jcb.140.1.49. J Cell Biol. 1998. PMID: 9425153 Free PMC article.
-
Peroxisomal membrane protein import does not require Pex17p.J Biol Chem. 2002 May 10;277(19):16498-504. doi: 10.1074/jbc.M111728200. Epub 2002 Feb 21. J Biol Chem. 2002. PMID: 11859077
-
Import of peroxisomal membrane proteins: the interplay of Pex3p- and Pex19p-mediated interactions.Biochim Biophys Acta. 2006 Dec;1763(12):1639-46. doi: 10.1016/j.bbamcr.2006.09.030. Epub 2006 Sep 26. Biochim Biophys Acta. 2006. PMID: 17069900 Review.
-
Peroxisome biogenesis disorders: molecular basis for impaired peroxisomal membrane assembly: in metabolic functions and biogenesis of peroxisomes in health and disease.Biochim Biophys Acta. 2012 Sep;1822(9):1337-42. doi: 10.1016/j.bbadis.2012.06.004. Epub 2012 Jun 13. Biochim Biophys Acta. 2012. PMID: 22705440 Review.
Cited by
-
Yarrowia lipolytica cells mutant for the peroxisomal peroxin Pex19p contain structures resembling wild-type peroxisomes.Mol Biol Cell. 2001 Nov;12(11):3353-64. doi: 10.1091/mbc.12.11.3353. Mol Biol Cell. 2001. PMID: 11694572 Free PMC article.
-
Ubiquitination of the peroxisomal import receptor Pex5p.Biochem J. 2004 Nov 15;384(Pt 1):37-45. doi: 10.1042/BJ20040572. Biochem J. 2004. PMID: 15283676 Free PMC article.
-
PEX3 gene knockout influences recombinant xylanase expression by Komagataella phaffii.Synth Syst Biotechnol. 2025 Mar 28;10(3):764-773. doi: 10.1016/j.synbio.2025.03.010. eCollection 2025 Sep. Synth Syst Biotechnol. 2025. PMID: 40248486 Free PMC article.
-
The peroxisome biogenesis factors pex4p, pex22p, pex1p, and pex6p act in the terminal steps of peroxisomal matrix protein import.Mol Cell Biol. 2000 Oct;20(20):7516-26. doi: 10.1128/MCB.20.20.7516-7526.2000. Mol Cell Biol. 2000. PMID: 11003648 Free PMC article.
-
Zellweger syndrome; identification of mutations in PEX19 and PEX26 gene in Saudi families.Ann Med. 2025 Dec;57(1):2447400. doi: 10.1080/07853890.2024.2447400. Epub 2025 Jan 6. Ann Med. 2025. PMID: 39757991 Free PMC article.
References
-
- Albertini M, Rehling P, Erdmann R, Girzalsky W, Kiel JA, Veenhuis M, Kunau WH. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell. 1997;89:83–92. - PubMed
-
- Baerends RJS, Rasmussen SW, Hilbrands RE, van der Heide M, Faber KN, Reuvekamp PTW, Kiel JAKW, Cregg JM, van der Klei I, Veenhuis M. The Hansenula polymorpha PER9 gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J Biol Chem. 1996;271:8887–8894. - PubMed
-
- Bodnar AG, Rachubinski RA. Characterization of the integral membrane polypeptides of rat liver peroxisomes isolated from untreated and clofibrate-treated rats. Biochem Cell Biol. 1991;69:499–508. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

