The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA
- PMID: 10588695
- PMCID: PMC24426
- DOI: 10.1073/pnas.96.25.14270
The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA
Abstract
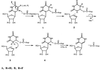
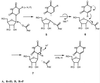
tRNA pseudouridine synthase I (PsiSI) catalyzes the conversion of uridine to Psi at positions 38, 39, and/or 40 in the anticodon loop of tRNAs. PsiSI forms a covalent adduct with 5-fluorouracil (FUra)-tRNA (tRNA(Phe) containing FUra in place of Ura) to form a putative analog of a steady-state intermediate in the normal reaction pathway. Previously, we proposed that a conserved aspartate of the enzyme serves as a nucleophilic catalyst in both the normal enzyme reaction and in the formation of a covalent complex with FUra-tRNA. The covalent adduct between FUra-tRNA and PsiSI was isolated and disrupted by hydrolysis and the FUra-tRNA was recovered. The target FU39 of the recovered FUra-tRNA was modified by the addition of water across the 5,6-double bond of the pyrimidine base to form 5,6-dihydro-6-hydroxy-5-fluorouridine. We deduced that the conserved aspartate of the enzyme adds to the 6-position of the target FUra to form a stable covalent adduct, which can undergo O-acyl hydrolytic cleavage to form the observed product. Assuming that an analogous covalent complex is formed in the normal reaction, we have deduced a complete mechanism for PsiS.
Figures






Similar articles
-
A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst.Biochemistry. 1998 Jan 6;37(1):344-51. doi: 10.1021/bi971874+. Biochemistry. 1998. PMID: 9425056
-
Major identity determinants for enzymatic formation of ribothymidine and pseudouridine in the T psi-loop of yeast tRNAs.J Mol Biol. 1997 Dec 12;274(4):505-18. doi: 10.1006/jmbi.1997.1417. J Mol Biol. 1997. PMID: 9417931
-
Interactions of transfer RNA pseudouridine synthases with RNAs substituted with fluorouracil.Nucleic Acids Res. 1991 Nov 25;19(22):6139-44. doi: 10.1093/nar/19.22.6139. Nucleic Acids Res. 1991. PMID: 1956773 Free PMC article.
-
Pseudouridine Modifications in Transfer RNA and tRNA Pseudouridine Synthases.J Mol Biol. 2025 Aug 15;437(16):169183. doi: 10.1016/j.jmb.2025.169183. Epub 2025 May 16. J Mol Biol. 2025. PMID: 40382211 Review.
-
Pseudouridines and pseudouridine synthases of the ribosome.Cold Spring Harb Symp Quant Biol. 2001;66:147-59. doi: 10.1101/sqb.2001.66.147. Cold Spring Harb Symp Quant Biol. 2001. PMID: 12762017 Review.
Cited by
-
Accurate placement of substrate RNA by Gar1 in H/ACA RNA-guided pseudouridylation.Nucleic Acids Res. 2015 Sep 3;43(15):7207-16. doi: 10.1093/nar/gkv757. Epub 2015 Jul 22. Nucleic Acids Res. 2015. PMID: 26206671 Free PMC article.
-
Pseudouridine: still mysterious, but never a fake (uridine)!RNA Biol. 2014;11(12):1540-54. doi: 10.4161/15476286.2014.992278. RNA Biol. 2014. PMID: 25616362 Free PMC article. Review.
-
Transglycosylation: a mechanism for RNA modification (and editing?).Bioorg Chem. 2005 Jun;33(3):229-51. doi: 10.1016/j.bioorg.2005.01.001. Epub 2005 Feb 23. Bioorg Chem. 2005. PMID: 15888313 Free PMC article. Review.
-
Mechanistic investigations of the pseudouridine synthase RluA using RNA containing 5-fluorouridine.Biochemistry. 2006 Oct 3;45(39):12029-38. doi: 10.1021/bi061293x. Biochemistry. 2006. PMID: 17002302 Free PMC article.
-
Unexpected linear ion trap collision-induced dissociation and Fourier transform ion cyclotron resonance infrared multi-photon dissociation fragmentation of a hydrated C-glycoside of 5-fluorouridine formed by the action of the pseudouridine synthases RluA and TruB.Rapid Commun Mass Spectrom. 2011 Sep 30;25(18):2627-32. doi: 10.1002/rcm.5162. Rapid Commun Mass Spectrom. 2011. PMID: 23657957 Free PMC article.
References
-
- Björk G R, Ericson J U, Gustafsson C E, Hagervall T G, Jönsson Y H, Wikström P M. Annu Rev Biochem. 1987;56:263–287. - PubMed
-
- Bakin A, Ofengand J. Biochemistry. 1993;32:9754–9762. - PubMed
-
- Santi D V, Wataya Y, Matsuda A. In: Proceedings of the International Symposium on Substrate-Induced Irreversible Inhibition of Enzymes. Seiler N, Jung M J, Koch-Weser J, editors. Strasbourg, France: Elsevier; 1978. pp. 291–303.
-
- Prior J J, Maley J, Santi D V. J Biol Chem. 1984;259:2422–2428. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

