The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome
- PMID: 10588719
- PMCID: PMC24450
- DOI: 10.1073/pnas.96.25.14412
The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome
Abstract
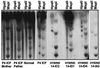
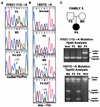
DNA methylation is an important regulator of genetic information in species ranging from bacteria to humans. DNA methylation appears to be critical for mammalian development because mice nullizygous for a targeted disruption of the DNMT1 DNA methyltransferase die at an early embryonic stage. No DNA methyltransferase mutations have been reported in humans until now. We describe here the first example of naturally occurring mutations in a mammalian DNA methyltransferase gene. These mutations occur in patients with a rare autosomal recessive disorder, which is termed the ICF syndrome, for immunodeficiency, centromeric instability, and facial anomalies. Centromeric instability of chromosomes 1, 9, and 16 is associated with abnormal hypomethylation of CpG sites in their pericentromeric satellite regions. We are able to complement this hypomethylation defect by somatic cell fusion to Chinese hamster ovary cells, suggesting that the ICF gene is conserved in the hamster and promotes de novo methylation. ICF has been localized to a 9-centimorgan region of chromosome 20 by homozygosity mapping. By searching for homologies to known DNA methyltransferases, we identified a genomic sequence in the ICF region that contains the homologue of the mouse Dnmt3b methyltransferase gene. Using the human sequence to screen ICF kindreds, we discovered mutations in four patients from three families. Mutations include two missense substitutions and a 3-aa insertion resulting from the creation of a novel 3' splice acceptor. None of the mutations were found in over 200 normal chromosomes. We conclude that mutations in the DNMT3B are responsible for the ICF syndrome.
Figures





References
-
- Tiepolo L, Maraschio P, Gimelli G, Cuoco C, Gargani G F, Romano C. Hum Genet. 1979;51:127–137. - PubMed
-
- Hulten M. Clin Genet. 1978;14:294.
-
- Jeanpierre M, Turleau C, Aurias A, Prieur M, Ledeist F, Fischer A, Viegas-Pequignot E. Hum Mol Genet. 1993;2:731–735. - PubMed
-
- Ji W, Hernandez R, Zhang X Y, Qu G Z, Frady A, Varela M, Ehrlich M. Mutat Res. 1997;379:33–41. - PubMed
-
- Miniou P, Jeanpierre M, Bourc’his D, Coutinho Barbosa A C, Blanquet V, Viegas-Pequignot E. Hum Genet. 1997;99:738–745. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

