Conserved characteristics of heterochromatin-forming DNA at the 15q11-q13 imprinting center
- PMID: 10588722
- PMCID: PMC24453
- DOI: 10.1073/pnas.96.25.14430
Conserved characteristics of heterochromatin-forming DNA at the 15q11-q13 imprinting center
Erratum in
- Proc Natl Acad Sci U S A 2000 Apr 11;97(8):4410
Abstract
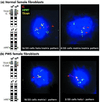
Nuclear matrix binding assays (NMBAs) define certain DNA sequences as matrix attachment regions (MARs), which often have cis-acting epigenetic regulatory functions. We used NMBAs to analyze the functionally important 15q11-q13 imprinting center (IC). We find that the IC is composed of an unusually high density of MARs, located in close proximity to the germ line elements that are proposed to direct imprint switching in this region. Moreover, we find that the organization of MARs is the same at the homologous mouse locus, despite extensive divergence of DNA sequence. MARs of this size are not usually associated with genes but rather with heterochromatin-forming areas of the genome. In contrast, the 15q11-q13 region contains multiple transcribed genes and is unusual for being subject to genomic imprinting, causing the maternal chromosome to be more transcriptionally silent, methylated, and late replicating than the paternal chromosome. We suggest that the extensive MAR sequences at the IC are organized as heterochromatin during oogenesis, an organization disrupted during spermatogenesis. Consistent with this model, multicolor fluorescence in situ hybridization to halo nuclei demonstrates a strong matrix association of the maternal IC, whereas the paternal IC is more decondensed, extending into the nuclear halo. This model also provides a mechanism for spreading of the imprinting signal, because heterochromatin at the IC on the maternal chromosome may exert a suppressive position effect in cis. We propose that the germ line elements at the 15q11-q13 IC mediate their effects through the candidate heterochromatin-forming DNA identified in this study.
Figures



Similar articles
-
Imprinting-mutation mechanisms in Prader-Willi syndrome.Am J Hum Genet. 1999 Feb;64(2):397-413. doi: 10.1086/302233. Am J Hum Genet. 1999. PMID: 9973278 Free PMC article.
-
Allele-specific expression analysis by RNA-FISH demonstrates preferential maternal expression of UBE3A and imprint maintenance within 15q11- q13 duplications.Hum Mol Genet. 2002 Jul 15;11(15):1707-18. doi: 10.1093/hmg/11.15.1707. Hum Mol Genet. 2002. PMID: 12095913
-
Minimal definition of the imprinting center and fixation of chromosome 15q11-q13 epigenotype by imprinting mutations.Proc Natl Acad Sci U S A. 1996 Jul 23;93(15):7811-5. doi: 10.1073/pnas.93.15.7811. Proc Natl Acad Sci U S A. 1996. PMID: 8755558 Free PMC article.
-
Mechanisms of imprinting of the Prader-Willi/Angelman region.Am J Med Genet A. 2008 Aug 15;146A(16):2041-52. doi: 10.1002/ajmg.a.32364. Am J Med Genet A. 2008. PMID: 18627066 Review.
-
Genomic imprinting: potential function and mechanisms revealed by the Prader-Willi and Angelman syndromes.Mol Hum Reprod. 1997 Apr;3(4):321-32. doi: 10.1093/molehr/3.4.321. Mol Hum Reprod. 1997. PMID: 9237260 Review.
Cited by
-
Phylogenetic footprint analysis of IGF2 in extant mammals.Genome Res. 2004 Sep;14(9):1726-32. doi: 10.1101/gr.2774804. Genome Res. 2004. PMID: 15342558 Free PMC article.
-
Transposable elements as scaffold/matrix attachment regions: shaping organization and functions in genomes.Front Mol Biosci. 2024 Feb 22;10:1326933. doi: 10.3389/fmolb.2023.1326933. eCollection 2023. Front Mol Biosci. 2024. PMID: 38455359 Free PMC article.
-
A new role for expressed pseudogenes as ncRNA: regulation of mRNA stability of its homologous coding gene.J Mol Med (Berl). 2004 Jul;82(7):414-22. doi: 10.1007/s00109-004-0550-3. Epub 2004 May 18. J Mol Med (Berl). 2004. PMID: 15148580
-
Regulatory elements associated with paternally-expressed genes in the imprinted murine Angelman/Prader-Willi syndrome domain.PLoS One. 2013;8(2):e52390. doi: 10.1371/journal.pone.0052390. Epub 2013 Feb 4. PLoS One. 2013. PMID: 23390487 Free PMC article.
-
The Drosophila homolog of the mammalian imprint regulator, CTCF, maintains the maternal genomic imprint in Drosophila melanogaster.BMC Biol. 2010 Jul 30;8:105. doi: 10.1186/1741-7007-8-105. BMC Biol. 2010. PMID: 20673338 Free PMC article.
References
-
- Nicholls R D, Saitoh S, Horsthemke B. Trends Genet. 1998;14:194–200. - PubMed
-
- Kitsberg D, Selig S, Brandeis M, Simon I, Keshet I, Driscoll D J, Nicholls R D, Cedar H. Nature (London) 1993;364:459–463. - PubMed
-
- Gunaratne P H, Nakao M, Ledbetter D H, Sutcliffe J S, Chinault A C. Genes Dev. 1995;9:808–820. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

