Functioning of the Drosophila integral U1/U2 protein Snf independent of U1 and U2 small nuclear ribonucleoprotein particles is revealed by snf(+) gene dose effects
- PMID: 10588726
- PMCID: PMC24457
- DOI: 10.1073/pnas.96.25.14451
Functioning of the Drosophila integral U1/U2 protein Snf independent of U1 and U2 small nuclear ribonucleoprotein particles is revealed by snf(+) gene dose effects
Abstract
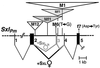
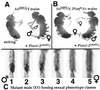
Snf, encoded by sans fille, is the Drosophila homolog of mammalian U1A and U2B" and is an integral component of U1 and U2 small nuclear ribonucleoprotein particles (snRNPs). Surprisingly, changes in the level of this housekeeping protein can specifically affect autoregulatory activity of the RNA-binding protein Sex-lethal (Sxl) in an action that we infer must be physically separate from Snf's functioning within snRNPs. Sxl is a master switch gene that controls its own pre-mRNA splicing as well as splicing for subordinate switch genes that regulate sex determination and dosage compensation. Exploiting an unusual new set of mutant Sxl alleles in an in vivo assay, we show that Snf is rate-limiting for Sxl autoregulation when Sxl levels are low. In such situations, increasing either maternal or zygotic snf(+) dose enhances the positive autoregulatory activity of Sxl for Sxl somatic pre-mRNA splicing without affecting Sxl activities toward its other RNA targets. In contrast, increasing the dose of genes encoding either the integral U1 snRNP protein U1-70k, or the integral U2 snRNP protein SF3a(60), has no effect. Increased snf(+) enhances Sxl autoregulation even when U1-70k and SF3a(60) are reduced by mutation to levels that, in the case of SF3a(60), demonstrably interfere with Sxl autoregulation. The observation that increased snf(+) does not suppress other phenotypes associated with mutations that reduce U1-70k or SF3a(60) is additional evidence that snf(+) dose effects are not caused by increased snRNP levels. Mammalian U1A protein, like Snf, has a snRNP-independent function.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

