Bacterial diversity within the human subgingival crevice
- PMID: 10588742
- PMCID: PMC24473
- DOI: 10.1073/pnas.96.25.14547
Bacterial diversity within the human subgingival crevice
Abstract
Molecular, sequence-based environmental surveys of microorganisms have revealed a large degree of previously uncharacterized diversity. However, nearly all studies of the human endogenous bacterial flora have relied on cultivation and biochemical characterization of the resident organisms. We used molecular methods to characterize the breadth of bacterial diversity within the human subgingival crevice by comparing 264 small subunit rDNA sequences from 21 clone libraries created with products amplified directly from subgingival plaque, with sequences obtained from bacteria that were cultivated from the same specimen, as well as with sequences available in public databases. The majority (52.5%) of the directly amplified 16S rRNA sequences were <99% identical to sequences within public databases. In contrast, only 21.4% of the sequences recovered from cultivated bacteria showed this degree of variability. The 16S rDNA sequences recovered by direct amplification were also more deeply divergent; 13.5% of the amplified sequences were more than 5% nonidentical to any known sequence, a level of dissimilarity that is often found between members of different genera. None of the cultivated sequences exhibited this degree of sequence dissimilarity. Finally, direct amplification of 16S rDNA yielded a more diverse view of the subgingival bacterial flora than did cultivation. Our data suggest that a significant proportion of the resident human bacterial flora remain poorly characterized, even within this well studied and familiar microbial environment.
Figures

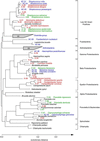

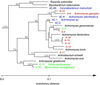
Similar articles
-
Bacterial diversity in human subgingival plaque.J Bacteriol. 2001 Jun;183(12):3770-83. doi: 10.1128/JB.183.12.3770-3783.2001. J Bacteriol. 2001. PMID: 11371542 Free PMC article.
-
Novel subgingival bacterial phylotypes detected using multiple universal polymerase chain reaction primer sets.Oral Microbiol Immunol. 2006 Feb;21(1):61-8. doi: 10.1111/j.1399-302X.2005.00255.x. Oral Microbiol Immunol. 2006. PMID: 16390343
-
Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens.Microbiology (Reading). 2003 Jan;149(Pt 1):67-75. doi: 10.1099/mic.0.25791-0. Microbiology (Reading). 2003. PMID: 12576581
-
Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease.Appl Environ Microbiol. 2003 Mar;69(3):1687-94. doi: 10.1128/AEM.69.3.1687-1694.2003. Appl Environ Microbiol. 2003. PMID: 12620860 Free PMC article.
-
Microbial diversity and interactions in subgingival biofilm communities.Front Oral Biol. 2012;15:17-40. doi: 10.1159/000329669. Epub 2011 Nov 11. Front Oral Biol. 2012. PMID: 22142955 Review.
Cited by
-
Effects of nicotine on the growth and protein expression of Porphyromonas gingivalis.J Microbiol. 2012 Feb;50(1):143-8. doi: 10.1007/s12275-012-1212-8. Epub 2012 Feb 27. J Microbiol. 2012. PMID: 22367949
-
High-quality draft genome sequences of five anaerobic oral bacteria and description of Peptoanaerobacter stomatis gen. nov., sp. nov., a new member of the family Peptostreptococcaceae.Stand Genomic Sci. 2015 Jul 18;10:37. doi: 10.1186/s40793-015-0027-8. eCollection 2015. Stand Genomic Sci. 2015. PMID: 26221418 Free PMC article.
-
Wound biofilms: lessons learned from oral biofilms.Wound Repair Regen. 2013 May-Jun;21(3):352-62. doi: 10.1111/wrr.12034. Epub 2013 Apr 1. Wound Repair Regen. 2013. PMID: 23551419 Free PMC article. Review.
-
Organo-selenium-containing dental sealant inhibits bacterial biofilm.J Dent Res. 2013 May;92(5):461-6. doi: 10.1177/0022034513482141. Epub 2013 Mar 8. J Dent Res. 2013. PMID: 23475900 Free PMC article.
-
The Hoops, Hopes, and Hypes of Human Microbiome Research.Yale J Biol Med. 2016 Sep 30;89(3):363-373. eCollection 2016 Sep. Yale J Biol Med. 2016. PMID: 27698620 Free PMC article. Review.
References
-
- Mackowiak P A. N Engl J Med. 1982;307:83–93. - PubMed
-
- Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Nature (London) 1990;345:60–63. - PubMed
-
- Stackebrandt E, Liesack W, Goebel B M. FASEB J. 1993;7:232–236. - PubMed
-
- Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkins L S. N Engl J Med. 1990;323:1573–1580. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

