The circadian clock controls the expression pattern of the circadian input photoreceptor, phytochrome B
- PMID: 10588760
- PMCID: PMC24491
- DOI: 10.1073/pnas.96.25.14652
The circadian clock controls the expression pattern of the circadian input photoreceptor, phytochrome B
Abstract
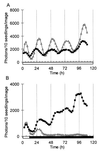
Developmental and physiological responses are regulated by light throughout the entire life cycle of higher plants. To sense changes in the light environment, plants have developed various photoreceptors, including the red/far-red light-absorbing phytochromes and blue light-absorbing cryptochromes. A wide variety of physiological responses, including most light responses, also are modulated by circadian rhythms that are generated by an endogenous oscillator, the circadian clock. To provide information on local time, circadian clocks are synchronized and entrained by environmental time cues, of which light is among the most important. Light-driven entrainment of the Arabidopsis circadian clock has been shown to be mediated by phytochrome A (phyA), phytochrome B (phyB), and cryptochromes 1 and 2, thus affirming the roles of these photoreceptors as input regulators to the plant circadian clock. Here we show that the expression of PHYB::LUC reporter genes containing the promoter and 5' untranslated region of the tobacco NtPHYB1 or Arabidopsis AtPHYB genes fused to the luciferase (LUC) gene exhibit robust circadian oscillations in transgenic plants. We demonstrate that the abundance of PHYB RNA retains this circadian regulation and use a PHYB::Luc fusion protein to show that the rate of PHYB synthesis is also rhythmic. The abundance of bulk PHYB protein, however, exhibits only weak circadian rhythmicity, if any. These data suggest that photoreceptor gene expression patterns may be significant in the daily regulation of plant physiology and indicate an unexpectedly intimate relationship between the components of the input pathway and the putative circadian clock mechanism in higher plants.
Figures




References
-
- Kendrick R E, Kronenberg G H M. Photomorphogenesis in Plants. Dordrecht, The Netherlands: Kluwer; 1994.
-
- Sharrock R A, Quail P H. Genes Dev. 1989;3:1745–1757. - PubMed
-
- Koornneef M, Rolff E, Spruit C J P. Z Pflanzenphysiol. 1980;100:147–160.
-
- Wester L, Somers D E, Clack T, Sharrock R A. Plant J. 1994;5:261–272. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

