Different regions of hepatitis B virus X protein are required for enhancement of bZip-mediated transactivation versus transrepression
- PMID: 10590094
- PMCID: PMC111516
- DOI: 10.1128/jvi.74.1.83-90.2000
Different regions of hepatitis B virus X protein are required for enhancement of bZip-mediated transactivation versus transrepression
Abstract
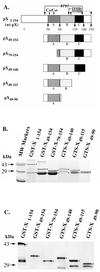
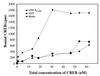
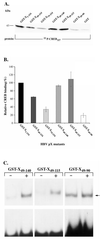
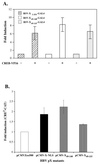
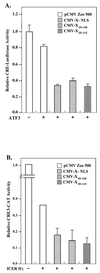
The hepatitis B virus X protein (pX) interacts directly with the bZip transactivator CREB and the bZip repressors ICERIIgamma and ATF3, increasing their DNA-binding affinity in vitro and their transcriptional efficacy in vivo. However, the mechanism of bZip-pX interaction and of the pX-mediated increase in the bZip transcriptional efficacy remains to be understood. In this study with deletion mutants of pX, we delineated a 67-amino-acid region spanning residues 49 to 115 required for direct CREB, ATF3, and ICER IIgamma interaction in vitro and in vivo and increased bZip/CRE binding in vitro. Transient transfections of the pX deletion mutants in AML12 hepatocytes demonstrate that pX(49-115) is as effective as the full-length pX in enhancing the ATF3- and ICERIIgamma-mediated transrepression. However, this pX region is inactive in increasing the transactivation efficacy of CREB; additional amino acid residues present in pX(49-140) are required to mediate the increased transactivation efficacy of CREB in vivo. This requirement for different regions of pX in affecting CREB transactivation suggests that amino acid residues 115 to 140 integrate additional events in effecting pX-mediated transactivation, such as concomitant interactions with select components of the basal transcriptional apparatus.
Figures


References
-
- Arii M, Takada K, Koike K. Identification of three essential regions of hepatitis B virus X protein for trans-activation function. Oncogene. 1992;7:397–403. - PubMed
-
- Barnabas S, Hai T, Andrisani O M. The hepatitis B virus X protein enhances the DNA-binding potential and transcription efficacy of bZip transcription factors. J Biol Chem. 1997;272:20684–20690. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

