Conditional site-specific integration into human chromosome 19 by using a ligand-dependent chimeric adeno-associated virus/Rep protein
- PMID: 10590116
- PMCID: PMC111538
- DOI: 10.1128/jvi.74.1.281-294.2000
Conditional site-specific integration into human chromosome 19 by using a ligand-dependent chimeric adeno-associated virus/Rep protein
Abstract
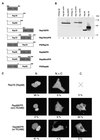
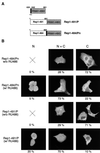
It is of great interest for gene therapy to develop vectors that drive the insertion of a therapeutic gene into a chosen specific site on the cellular genome. Adeno-associated virus (AAV) is unique among mammalian viruses in that it integrates into a distinct region of human chromosome 19 (integration site AAVS1). The inverted terminal repeats (ITRs) flanking the AAV genome and the AAV-encoded nonstructural proteins Rep78 and/or Rep68 are the only viral elements necessary and sufficient for site-specific integration. However, it is also known that unrestrained Rep activity may cause nonspecific genomic rearrangements at AAVS1 and/or have detrimental effects on cell physiology. In this paper we describe the generation of a ligand-dependent form of Rep, obtained by fusing a C-terminally deleted Rep68 with a truncated form of the hormone binding domain of the human progesterone receptor, which does not bind progesterone but binds only its synthetic antagonist RU486. The activity of this chimeric protein, named Rep1-491/P, is highly dependent on RU486 in various assays: in particular, it triggers site-specific integration at AAVS1 of an ITR-flanked cassette in a ligand-dependent manner, as efficiently as wild-type Rep68 but without generating unwanted genomic rearrangement at AAVS1.
Figures





Similar articles
-
Herpes simplex virus type 1/adeno-associated virus hybrid vectors mediate site-specific integration at the adeno-associated virus preintegration site, AAVS1, on human chromosome 19.J Virol. 2002 Jul;76(14):7163-73. doi: 10.1128/jvi.76.14.7163-7173.2002. J Virol. 2002. PMID: 12072516 Free PMC article.
-
Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome.J Virol. 1997 Oct;71(10):7951-9. doi: 10.1128/JVI.71.10.7951-7959.1997. J Virol. 1997. PMID: 9311886 Free PMC article.
-
Roles of adeno-associated virus Rep protein and human chromosome 19 in site-specific recombination.J Virol. 2000 May;74(9):3953-66. doi: 10.1128/jvi.74.9.3953-3966.2000. J Virol. 2000. PMID: 10756007 Free PMC article.
-
Site-specific integration by the adeno-associated virus rep protein.Curr Gene Ther. 2011 Oct;11(5):399-405. doi: 10.2174/156652311797415809. Curr Gene Ther. 2011. PMID: 21827397 Review.
-
Site-specific integration by adeno-associated virus.Proc Natl Acad Sci U S A. 1996 Oct 15;93(21):11288-94. doi: 10.1073/pnas.93.21.11288. Proc Natl Acad Sci U S A. 1996. PMID: 8876128 Free PMC article. Review.
Cited by
-
Regulated gene insertion by steroid-induced PhiC31 integrase.Nucleic Acids Res. 2008 Jun;36(11):e67. doi: 10.1093/nar/gkn298. Epub 2008 May 22. Nucleic Acids Res. 2008. PMID: 18499713 Free PMC article.
-
Gene Therapy for Cardiovascular Disease.J Biomed Biotechnol. 2003;2003(2):138-148. doi: 10.1155/S1110724303209086. J Biomed Biotechnol. 2003. PMID: 12721517 Free PMC article.
-
Targeted chromosomal insertion of large DNA into the human genome by a fiber-modified high-capacity adenovirus-based vector system.PLoS One. 2008 Aug 29;3(8):e3084. doi: 10.1371/journal.pone.0003084. PLoS One. 2008. PMID: 18769728 Free PMC article.
-
Stimulation of homology-directed gene targeting at an endogenous human locus by a nicking endonuclease.Nucleic Acids Res. 2009 Sep;37(17):5725-36. doi: 10.1093/nar/gkp643. Epub 2009 Aug 3. Nucleic Acids Res. 2009. PMID: 19651880 Free PMC article.
-
Adeno-associated virus (AAV) site-specific recombination does not require a Rep-dependent origin of replication within the AAV terminal repeat.Proc Natl Acad Sci U S A. 2001 Nov 20;98(24):13525-30. doi: 10.1073/pnas.241508998. Epub 2001 Nov 13. Proc Natl Acad Sci U S A. 2001. PMID: 11707592 Free PMC article.
References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995.
-
- Baulieu E E. RU486: an antiprogestin steroid with contragestive activity in women. In: Baulieu E E, Segal J, editors. The antiprogestin steroid RU486 and human fertility control. New York, N.Y: Plenum Press; 1985. pp. 1–25.
-
- Berns K I, Linden R M. The cryptic life stile of adeno-associated virus. Bioessays. 1995;17:237–245. - PubMed
-
- Cadepond F, Ulmann A, Baulieu E-E. RU486 (mifepristone): mechanisms of action and clinical use. Annu Rev Med. 1997;48:129–156. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

