Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal
- PMID: 10590135
- PMCID: PMC111557
- DOI: 10.1128/jvi.74.1.456-464.2000
Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal
Abstract
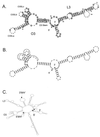
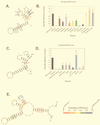
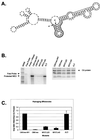
We previously identified a 160-nucleotide packaging signal, MPsi, from the 5' end of the Rous sarcoma virus genome. In this study, we determine the secondary structure of MPsi by using phylogenetic analysis with computer modeling and heterologous packaging assays of point mutants. The results of the in vivo studies are in good agreement with the computer model. Additionally, the packaging studies indicate several structures which are important for efficient packaging, including a single-stranded bulge containing the initiation codon for the short open reading frame, uORF3, as well as adjacent stem structures. Finally, we show that the L3 stem-loop at the 3' end of MPsi is dispensable for packaging, thus identifying an 82-nucleotide minimal packaging signal, microPsi, composed of the O3 stem-loop.
Figures


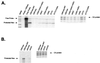
References
-
- Banks J D, Beemon K L, Linial M L. RNA regulatory elements in the genomes of simple retroviruses. Semin Virol. 1997;8:194–204.
-
- Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

