Identification of a bovine coronavirus packaging signal
- PMID: 10590153
- PMCID: PMC111575
- DOI: 10.1128/jvi.74.1.580-583.2000
Identification of a bovine coronavirus packaging signal
Abstract
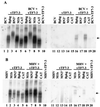
A region of the bovine coronavirus (BCV) genome that functions as a packaging signal has been cloned. The 291-nucleotide clone shares 72% homology with the region of mouse hepatitis coronavirus (MHV) gene 1b that contains the packaging signal. RNA transcripts were packaged into both BCV and MHV virions when the cloned region was appended to a noncoronavirus RNA. This is the first identification of a BCV packaging signal. The data demonstrate that the BCV genome contains a sequence that is conserved at both the sequence and functional levels, thus broadening our insight into coronavirus packaging.
Figures



References
-
- Bredenbeek B J, Pachuk C J, Notern A F H, Charite J, Luytjes W, Weiss S R, Spaan W J M. The primary structure and expression of the 2 open reading frame of the polymerase gene of the coronavirus MHV-A59—a highly conserved polymerase is expressed by an efficient ribosomal frame-shifting mechanism. Nucleic Acids Res. 1990;18:1825–1832. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

