Species specificity of macaque rhadinovirus glycoprotein B sequences
- PMID: 10590154
- PMCID: PMC111576
- DOI: 10.1128/jvi.74.1.584-590.2000
Species specificity of macaque rhadinovirus glycoprotein B sequences
Abstract
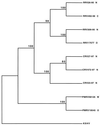
All members of the Herpesviridae family contain sequences for a highly conserved glycoprotein B (gB) gene. We investigated the phylogenetic relationships of gB sequences from eight independent rhadinovirus isolates obtained from three species: rhesus (Macaca mulatta), cynomologus (Macaca fasicularis), and pig-tailed (Macaca nemestrina) macaques. Samples were derived from monkeys housed at four separate facilities. Analysis of these eight independent gB sequences revealed five regions of heterogeneity within the 823- to 829-amino-acid polypeptides: residues 1 to 65, 120 to 185, 255 to 300, 352 to 393, and 412 to 457. The remaining regions of gB were highly conserved among the different macaque isolates. Overall divergence among these gene sequences ranged from 0.1 to 7.2% at the amino acid level. Phylogenetic trees constructed with our macaque rhadinovirus gB sequences and those derived from additional subfamilies or genera (alpha, beta, gamma-1, and gamma-2) revealed that the macaque gB sequences branched with other gamma-2 herpesvirus gB sequences and that within the gamma-2 genera, the macaque gB sequences clustered as a distinct branch. The eight macaque rhadinovirus gB sequences were all approximately equidistant from Kaposi sarcoma-associated herpesvirus (KSHV) gB sequences and had a shorter evolutionary distance to KSHV gB sequences than to any other herpesvirus, including the gamma-2 herpesvirus saimiri (HVS) of New World squirrel monkeys. The macaque gB sequences did not cluster according to the facility of origin, but did cluster according to the species of origin, displaying less intraspecies divergence (0.1 to 2.9%) than interspecies divergence (3.3 to 7.2%). These results demonstrate a close relatedness of rhadinovirus isolates from different macaque species.
Figures



References
-
- Alexander S, Elder J H. Carbohydrate dramatically influences immune reactivity of antisera to viral glycoprotein antigens. Science. 1984;226:1328–1330. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

