Mouse tumor model for neurofibromatosis type 1
- PMID: 10591653
- PMCID: PMC3079436
- DOI: 10.1126/science.286.5447.2176
Mouse tumor model for neurofibromatosis type 1
Abstract
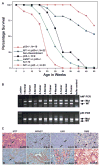
Neurofibromatosis type 1 (NF1) is an autosomal dominant disorder characterized by increased incidence of benign and malignant tumors of neural crest origin. Mutations that activate the protooncogene ras, such as loss of Nf1, cooperate with inactivating mutations at the p53 tumor suppressor gene during malignant transformation. One hundred percent of mice harboring null Nf1 and p53 alleles in cis synergize to develop soft tissue sarcomas between 3 and 7 months of age. These sarcomas exhibit loss of heterozygosity at both gene loci and express phenotypic traits characteristic of neural crest derivatives and human NF1 malignancies.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

