Increased coverage of protein families with the blocks database servers
- PMID: 10592233
- PMCID: PMC102407
- DOI: 10.1093/nar/28.1.228
Increased coverage of protein families with the blocks database servers
Abstract
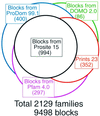
The Blocks Database WWW (http://blocks.fhcrc.org ) and Email (blocks@blocks.fhcrc.org ) servers provide tools to search DNA and protein queries against the Blocks+ Database of multiple alignments, which represent conserved protein regions. Blocks+ nearly doubles the number of protein families included in the database by adding families from the Pfam-A, ProDom and Domo databases to those from PROSITE and PRINTS. Other new features include improved Block Searcher statistics, searching with NCBI's IMPALA program and 3D display of blocks on PDB structures.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

