Histone H1 is dispensable for methylation-associated gene silencing in Ascobolus immersus and essential for long life span
- PMID: 10594009
- PMCID: PMC85047
- DOI: 10.1128/MCB.20.1.61-69.2000
Histone H1 is dispensable for methylation-associated gene silencing in Ascobolus immersus and essential for long life span
Abstract
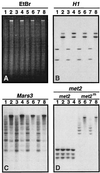
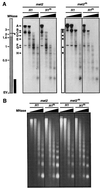
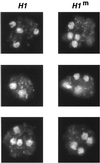
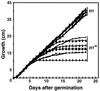
A gene encoding a protein that shows sequence similarity with the histone H1 family only was cloned in Ascobolus immersus. The deduced peptide sequence presents the characteristic three-domain structure of metazoan linker histones, with a central globular region, an N-terminal tail, and a long positively charged C-terminal tail. By constructing an artificial duplication of this gene, named H1, it was possible to methylate and silence it by the MIP (methylation induced premeiotically) process. This resulted in the complete loss of the Ascobolus H1 histone. Mutant strains lacking H1 displayed normal methylation-associated gene silencing, underwent MIP, and showed the same methylation-associated chromatin modifications as did wild-type strains. However, they displayed an increased accessibility of micrococcal nuclease to chromatin, whether DNA was methylated or not, and exhibited a hypermethylation of the methylated genome compartment. These features are taken to imply that Ascobolus H1 histone is a ubiquitous component of chromatin which plays no role in methylation-associated gene silencing. Mutant strains lacking histone H1 reproduced normally through sexual crosses and displayed normal early vegetative growth. However, between 6 and 13 days after germination, they abruptly and consistently stopped growing, indicating that Ascobolus H1 histone is necessary for long life span. This constitutes the first observation of a physiologically important phenotype associated with the loss of H1.
Figures



References
-
- Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol. 1986;187:591–601. - PubMed
-
- Antequera F, Boyes J, Bird A P. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;62:503–514. - PubMed
-
- Bouvet P, Dimitrov S, Wolffe A P. Specific regulation of Xenopus chromosomal 5S rRNA gene transcription in vivo by histone H1. Genes Dev. 1994;8:1147–1159. - PubMed
-
- Campoy F J, Meehan R R, McKay S, Nixon J, Bird A P. Binding of histone H1 to DNA is indifferent to methylation at CpG sequences. J Biol Chem. 1995;270:26473–26481. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
