Auto-inhibition and partner proteins, core-binding factor beta (CBFbeta) and Ets-1, modulate DNA binding by CBFalpha2 (AML1)
- PMID: 10594012
- PMCID: PMC85059
- DOI: 10.1128/MCB.20.1.91-103.2000
Auto-inhibition and partner proteins, core-binding factor beta (CBFbeta) and Ets-1, modulate DNA binding by CBFalpha2 (AML1)
Abstract
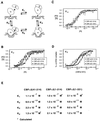
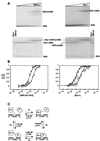
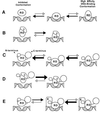
Core-binding factor alpha2 (CBFalpha2; otherwise known as AML1 or PEBP2alphaB) is a DNA-binding subunit in the family of core-binding factors (CBFs), heterodimeric transcription factors that play pivotal roles in multiple developmental processes in mammals, including hematopoiesis and bone development. The Runt domain in CBFalpha2 (amino acids 51 to 178) mediates DNA binding and heterodimerization with the non-DNA-binding CBFbeta subunit. Both the CBFbeta subunit and the DNA-binding protein Ets-1 stimulate DNA binding by the CBFalpha2 protein. Here we quantify and compare the extent of cooperativity between CBFalpha2, CBFbeta, and Ets-1. We also identify auto-inhibitory sequences within CBFalpha2 and sequences that modulate its interactions with CBFbeta and Ets-1. We show that sequences in the CBFalpha2 Runt domain and sequences C terminal to amino acid 214 inhibit DNA binding. Sequences C terminal to amino acid 214 also inhibit heterodimerization with the non-DNA-binding CBFbeta subunit, particularly heterodimerization off DNA. CBFbeta rescinds the intramolecular inhibition of CBFalpha2, stimulating DNA binding approximately 40-fold. In comparison, Ets-1 stimulates CBFalpha2 DNA binding 7- to 10-fold. Although the Runt domain alone is sufficient for heterodimerization with CBFbeta, sequences N terminal to amino acid 41 and between amino acids 190 and 214 are required for cooperative DNA binding with Ets-1. Cooperative DNA binding with Ets-1 is less pronounced with the CBFalpha2-CBFbeta heterodimer than with CBFalpha2 alone. These analyses demonstrate that CBFalpha2 is subject to both negative regulation by intramolecular interactions, and positive regulation by two alternative partnerships.
Figures





References
-
- Albagli O, Klaes A, Ferreira E, Leprince D, Klambt C. Function of ets genes is conserved between vertebrates and Drosophila. Mech Dev. 1996;59:29–40. - PubMed
-
- Bae S-C, Takahashi E, Zhang Y W, Ogawa E, Shigesada K, Namba Y, Satake M, Ito Y. Cloning, mapping and expression of PEBP2αC, a third gene encoding the mammalian Runt domain. Gene. 1995;159:245–248. - PubMed
-
- Bae S C, Yamaguchi-Iwai Y, Ogawa E, Maruyama M, Inuzuka M, Kagoshima H, Shigesada K, Satake M, Ito Y. Isolation of PEBP2αB cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene. 1993;8:809–814. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
