Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1alpha
- PMID: 10594042
- PMCID: PMC85095
- DOI: 10.1128/MCB.20.1.402-415.2000
Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1alpha
Abstract
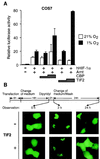
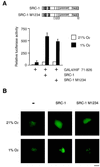
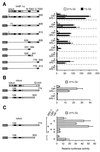
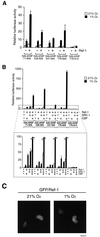
Hypoxia-inducible factor 1alpha (HIF-1alpha) functions as a transcription factor that is activated by decreased cellular oxygen concentrations to induce expression of a network of genes involved in angiogenesis, erythropoiesis, and glucose homeostasis. Here we demonstrate that two members of the SRC-1/p160 family of transcriptional coactivators harboring histone acetyltransferase activity, SRC-1 and transcription intermediary factor 2 (TIF2), are able to interact with HIF-1alpha and enhance its transactivation potential in a hypoxia-dependent manner. HIF-1alpha contains within its C terminus two transactivation domains. The hypoxia-inducible activity of both these domains was enhanced by either SRC-1 or the CREB-binding protein (CBP)/p300 coactivator. Moreover, at limiting concentrations, SRC-1 produced this effect in synergy with CBP. Interestingly, this effect was strongly potentiated by the redox regulatory protein Ref-1, a dual-function protein harboring DNA repair endonuclease and cysteine reducing activities. These data indicate that all three proteins, CBP, SRC-1, and Ref-1, are important components of the hypoxia signaling pathway and have a common function in regulation of HIF-1alpha function in hypoxic cells. Given the absence of cysteine residues in one of the Ref-1-regulated transactivation domains of HIF-1alpha, it is thus possible that Ref-1 functions in hypoxic cells by targeting critical steps in the recruitment of the CBP-SRC-1 coactivator complex.
Figures


References
-
- Antonsson C, Arulampalam V, Whitelaw M L, Pettersson S, Poellinger L. Constitutive function of the basic helix-loop-helix/PAS factor Arnt. Regulation of target promoters via the E box motif. J Biol Chem. 1995;270:13968–13972. - PubMed
-
- Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X-Y, Sauter G, Kallioniemi O-P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. - PubMed
-
- Arany Z, Sellers W R, Livingston D M, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77:799–800. - PubMed
-
- Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
