Characterization of a novel, antifungal, chitin-binding protein from Streptomyces tendae Tü901 that interferes with growth polarity
- PMID: 10601197
- PMCID: PMC94197
- DOI: 10.1128/JB.181.24.7421-7429.1999
Characterization of a novel, antifungal, chitin-binding protein from Streptomyces tendae Tü901 that interferes with growth polarity
Abstract
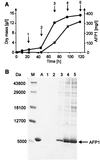
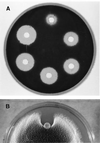
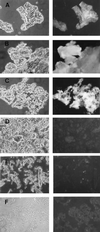
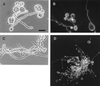
The afp1 gene, which encodes the antifungal protein AFP1, was cloned from nikkomycin-producing Streptomyces tendae Tü901, using a nikkomycin-negative mutant as a host and screening transformants for antifungal activity against Paecilomyces variotii in agar diffusion assays. The 384-bp afp1 gene has a low G+C content (63%) and a transcription termination structure with a poly(T) region, unusual attributes for Streptomyces genes. AFP1 was purified from culture filtrate of S. tendae carrying the afp1 gene on the multicopy plasmid pIJ699. The purified protein had a molecular mass of 9,862 Da and lacked a 42-residue N-terminal peptide deduced from the nucleotide sequence. AFP1 was stable at extreme pH values and high temperatures and toward commercial proteinases. AFP1 had limited similarity to cellulose-binding domains of microbial plant cell wall hydrolases and bound to crab shell chitin, chitosan, and cell walls of P. variotii but showed no enzyme activity. The biological activity of AFP1, which represents the first chitin-binding protein from bacteria exhibiting antifungal activity, was directed against specific ascomycetes, and synergistic interaction with the chitin synthetase inhibitor nikkomycin inhibited growth of Aspergillus species. Microscopy studies revealed that fluorescein-labeled AFP1 strongly bound to the surface of germinated conidia and to tips of growing hyphae, causing severe alterations in cell morphogenesis that gave rise to large spherical conidia and/or swollen hyphae and to atypical branching.
Figures

References
-
- Abad L R, D'Urzo M P, Liu D, Narasimhan M L, Reuveni M, Zhu J K, Niu X, K. S N, Hasegawa P M, Bressan R A. Antifungal activity of tobacco osmotin has specificity and involves plasma membrane permeabilization. Plant Sci. 1996;118:11–23.
-
- Bormann C, Aberle K, Fiedler H-P, Schrempf H. Genetic complementation of Streptomyces tendaedeficient in nikkomycin production. Appl Microbiol Biotechnol. 1990;32:424–430. - PubMed
-
- Bormann C, Mattern S, Schrempf H, Fiedler H P, Zähner H. Isolation of Streptomyces tendaemutants with an altered nikkomycin spectrum. J Antibiot. 1989;42:913–918. - PubMed
-
- Broekaert W F, Marien W, Terras F R G, De Brolle M F C, Proost P, Van Damme J, Dillen L, Claeys M, Rees S B, Vanderleyden J, Cammue B P A. Antimicrobial peptides from Amaranthus caudatusseeds with sequence homology to the cysteine/glycine-rich domain of chitin-binding proteins. Biochemistry. 1992;31:4308–4314. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

