Molecular analysis of the Candida albicans homolog of Saccharomyces cerevisiae MNN9, required for glycosylation of cell wall mannoproteins
- PMID: 10601199
- PMCID: PMC94199
- DOI: 10.1128/JB.181.24.7439-7448.1999
Molecular analysis of the Candida albicans homolog of Saccharomyces cerevisiae MNN9, required for glycosylation of cell wall mannoproteins
Abstract
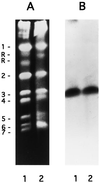
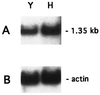
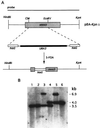
The fungal cell wall has generated interest as a potential target for developing antifungal drugs, and the genes encoding glucan and chitin in fungal pathogens have been studied to this end. Mannoproteins, the third major component of the cell wall, contain mannose in either O- or N-glycosidic linkages. Here we describe the molecular analysis of the Candida albicans homolog of Saccharomyces cerevisiae MNN9, a gene required for the synthesis of N-linked outer-chain mannan in yeast, and the phenotypes associated with its disruption. CaMNN9 has significant homology with S. cerevisiae MNN9, including a putative N-terminal transmembrane domain, and represents a member of a similar gene family in Candida. CaMNN9 resides on chromosome 3 and is expressed at similar levels in both yeast and hyphal cells. Disruption of both copies of CaMNN9 leads to phenotypic effects characteristic of cell wall defects including poor growth in liquid media and on solid media, formation of aggregates in liquid culture, osmotic sensitivity, aberrant hyphal formation, and increased sensitivity to lysis after treatment with beta-1,3-glucanase. Like all members of the S. cerevisiae MNN9 gene family the Camnn9Delta strain is resistant to sodium orthovanadate and sensitive to hygromycin B. Analysis of cell wall-associated carbohydrates showed the Camnn9Delta strain to contain half the amount of mannan present in cell walls derived from the wild-type parent strain. Reverse transcription-PCR and Northern analysis of the expression of MNN9 gene family members CaVAN1 and CaANP1 in the Camnn9Delta strain showed that transcription of those genes is not affected in the absence of CaMNN9 transcription. Our results suggest that, while the role MNN9 plays in glycosylation in both Candida and Saccharomyces is conserved, loss of MNN9 function in C. albicans leads to phenotypes that are inconsistent with the pathogenicity of the organism and thus identify CaMnn9p as a potential drug target.
Figures




Similar articles
-
Isolation of the MNN9 gene of Yarrowia lipolytica (YlMNN9) and phenotype analysis of a mutant ylmnn9 Delta strain.Yeast. 2003 May;20(7):633-44. doi: 10.1002/yea.990. Yeast. 2003. PMID: 12734801
-
Cloning and characterization of the Hansenula polymorpha homologue of the Saccharomyces cerevisiae MNN9 gene.Yeast. 2001 Mar 30;18(5):455-61. doi: 10.1002/yea.699. Yeast. 2001. PMID: 11255253
-
Sodium orthovanadate-resistant mutants of Saccharomyces cerevisiae show defects in Golgi-mediated protein glycosylation, sporulation and detergent resistance.Genetics. 1995 Jul;140(3):933-43. doi: 10.1093/genetics/140.3.933. Genetics. 1995. PMID: 7672592 Free PMC article.
-
N-glycosylation of yeast, with emphasis on Candida albicans.Med Mycol. 2001;39 Suppl 1:75-86. Med Mycol. 2001. PMID: 11800271 Review.
-
The Gas1 glycoprotein, a putative wall polymer cross-linker.Biochim Biophys Acta. 1999 Jan 6;1426(2):385-400. doi: 10.1016/s0304-4165(98)00138-x. Biochim Biophys Acta. 1999. PMID: 9878845 Review.
Cited by
-
Adaptations of the Secretome of Candida albicans in Response to Host-Related Environmental Conditions.Eukaryot Cell. 2015 Dec;14(12):1165-72. doi: 10.1128/EC.00142-15. Epub 2015 Oct 9. Eukaryot Cell. 2015. PMID: 26453650 Free PMC article. Review.
-
Surface stress induces a conserved cell wall stress response in the pathogenic fungus Candida albicans.Eukaryot Cell. 2013 Feb;12(2):254-64. doi: 10.1128/EC.00278-12. Epub 2012 Dec 14. Eukaryot Cell. 2013. PMID: 23243062 Free PMC article.
-
MNN5 encodes an iron-regulated alpha-1,2-mannosyltransferase important for protein glycosylation, cell wall integrity, morphogenesis, and virulence in Candida albicans.Eukaryot Cell. 2006 Feb;5(2):238-47. doi: 10.1128/EC.5.2.238-247.2006. Eukaryot Cell. 2006. PMID: 16467465 Free PMC article.
-
Role of Protein Glycosylation in Interactions of Medically Relevant Fungi with the Host.J Fungi (Basel). 2021 Oct 18;7(10):875. doi: 10.3390/jof7100875. J Fungi (Basel). 2021. PMID: 34682296 Free PMC article. Review.
-
Endoplasmic reticulum alpha-glycosidases of Candida albicans are required for N glycosylation, cell wall integrity, and normal host-fungus interaction.Eukaryot Cell. 2007 Dec;6(12):2184-93. doi: 10.1128/EC.00350-07. Epub 2007 Oct 12. Eukaryot Cell. 2007. PMID: 17933909 Free PMC article.
References
-
- Ballou C E. Yeast cell wall and cell surface. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces. Metabolism and gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. pp. 335–360.
-
- Ballou C E. Isolation, characterization and properties of Saccharomyces mnn mutants with non-conditional protein glycosylation defects. Methods Enzymol. 1990;185:440–476. - PubMed
-
- Ballou L, Cohen R E, Ballou C E. Saccharomyces cerevisiae mannoprotein mutants that make mannoproteins with a truncated outer chain. J Biol Chem. 1980;255:5986–5991. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

