Gradual alterations in cell wall structure and metabolism in vancomycin-resistant mutants of Staphylococcus aureus
- PMID: 10601215
- PMCID: PMC94215
- DOI: 10.1128/JB.181.24.7566-7570.1999
Gradual alterations in cell wall structure and metabolism in vancomycin-resistant mutants of Staphylococcus aureus
Abstract
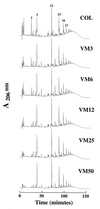
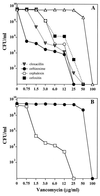
In five vancomycin-resistant laboratory step mutants selected from the highly and homogeneously methicillin-resistant Staphylococcus aureus strain COL (MIC of methicillin, 800 microg/ml; MIC of vancomycin, 1.5 microg/ml), the gradually increasing levels of resistance to vancomycin were accompanied by parallel decreases in the levels of methicillin resistance and abnormalities in cell wall metabolism. The latter included a gradual reduction in the proportion of highly cross-linked muropeptide species in peptidoglycan, down-regulation of the production of penicillin-binding protein 2A (PBP2A) and PBP4, and hypersensitivity to beta-lactam antibiotics each with a relatively selective affinity for the various staphylococcal PBPs; the PBP2-specific inhibitor ceftizoxime was particularly effective.
Figures


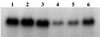

Similar articles
-
Recruitment of the mecA gene homologue of Staphylococcus sciuri into a resistance determinant and expression of the resistant phenotype in Staphylococcus aureus.J Bacteriol. 2001 Apr;183(8):2417-24. doi: 10.1128/JB.183.8.2417-2424.2001. J Bacteriol. 2001. PMID: 11274099 Free PMC article.
-
Moenomycin-resistance is associated with vancomycin-intermediate susceptibility in Staphylococcus aureus.Microbiol Immunol. 2003;47(12):927-35. doi: 10.1111/j.1348-0421.2003.tb03466.x. Microbiol Immunol. 2003. PMID: 14695442
-
Abnormal peptidoglycan produced in a methicillin-resistant strain of Staphylococcus aureus grown in the presence of methicillin: functional role for penicillin-binding protein 2A in cell wall synthesis.Antimicrob Agents Chemother. 1993 Feb;37(2):342-6. doi: 10.1128/AAC.37.2.342. Antimicrob Agents Chemother. 1993. PMID: 8452368 Free PMC article.
-
[The mechanism of vancomycin-resistant in Staphylococcus aureus].Tanpakushitsu Kakusan Koso. 2000 Jun;45(8):1329-38. Tanpakushitsu Kakusan Koso. 2000. PMID: 10846471 Review. Japanese. No abstract available.
-
Factors influencing methicillin resistance in staphylococci.Arch Microbiol. 2002 Sep;178(3):165-71. doi: 10.1007/s00203-002-0436-0. Epub 2002 Jun 19. Arch Microbiol. 2002. PMID: 12189417 Review.
Cited by
-
Mechanisms of resistance and clinical relevance of resistance to β-lactams, glycopeptides, and fluoroquinolones.Mayo Clin Proc. 2012 Feb;87(2):198-208. doi: 10.1016/j.mayocp.2011.12.003. Mayo Clin Proc. 2012. PMID: 22305032 Free PMC article. Review.
-
Generation of ramoplanin-resistant Staphylococcus aureus.FEMS Microbiol Lett. 2010 Sep 1;310(2):104-11. doi: 10.1111/j.1574-6968.2010.02051.x. Epub 2010 Jul 2. FEMS Microbiol Lett. 2010. PMID: 20659164 Free PMC article.
-
Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications.Clin Microbiol Rev. 2010 Jan;23(1):99-139. doi: 10.1128/CMR.00042-09. Clin Microbiol Rev. 2010. PMID: 20065327 Free PMC article. Review.
-
Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus.Proc Natl Acad Sci U S A. 2010 Nov 2;107(44):18991-6. doi: 10.1073/pnas.1004304107. Epub 2010 Oct 13. Proc Natl Acad Sci U S A. 2010. PMID: 20944066 Free PMC article.
-
An elevated mutation frequency favors development of vancomycin resistance in Staphylococcus aureus.Antimicrob Agents Chemother. 2002 Nov;46(11):3540-8. doi: 10.1128/AAC.46.11.3540-3548.2002. Antimicrob Agents Chemother. 2002. PMID: 12384362 Free PMC article.
References
-
- De Jonge B L M, Chang Y-S, Gage D, Tomasz A. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain: the role of penicillin binding protein 2A. J Biol Chem. 1992;267:11248–11254. - PubMed
-
- Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. - PubMed
-
- Murakami K, Fujimura T, Doi M. Nucleotide sequence of the structural gene for the penicillin-binding protein 2 of Staphylococcus aureus and the presence of a homologous gene in other staphylococci. FEMS Microbiol Lett. 1994;117:131–136. - PubMed
-
- Okonogi K, Noji Y, Nakao M, Imada A. The possible physiological roles of penicillin-binding proteins of methicillin-susceptible and methicillin-resistant Staphylococcus aureus. J Infect Chemother. 1995;1:50–58.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

