IS1630 of Mycoplasma fermentans, a novel IS30-type insertion element that targets and duplicates inverted repeats of variable length and sequence during insertion
- PMID: 10601219
- PMCID: PMC94219
- DOI: 10.1128/JB.181.24.7597-7607.1999
IS1630 of Mycoplasma fermentans, a novel IS30-type insertion element that targets and duplicates inverted repeats of variable length and sequence during insertion
Abstract
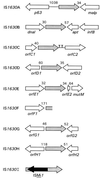
A new insertion sequence (IS) of Mycoplasma fermentans is described. This element, designated IS1630, is 1,377 bp long and has 27-bp inverted repeats at the termini. A single open reading frame (ORF), predicted to encode a basic protein of either 366 or 387 amino acids (depending on the start codon utilized), occupies most of this compact element. The predicted translation product of this ORF has homology to transposases of the IS30 family of IS elements and is most closely related (27% identical amino acid residues) to the product of the prototype of the group, IS30. Multiple copies of IS1630 are present in the genomes of at least two M. fermentans strains. Characterization and comparison of nine copies of the element revealed that IS1630 exhibits unusual target site specificity and, upon insertion, duplicates target sequences in a manner unlike that of any other IS element. IS1630 was shown to have the striking ability to target and duplicate inverted repeats of variable length and sequence during transposition. IS30-type elements typically generate 2- or 3-bp target site duplications, whereas those created by IS1630 vary between 19 and 26 bp. With the exception of two recently reported IS4-type elements which have the ability to generate variable large duplications (B. B. Plikaytis, J. T. Crawford, and T. M. Shinnick, J. Bacteriol. 180:1037-1043, 1998; E. M. Vilei, J. Nicolet, and J. Frey, J. Bacteriol. 181:1319-1323, 1999), such large direct repeats had not been observed for other IS elements. Interestingly, the IS1630-generated duplications are all symmetrical inverted repeat sequences that are apparently derived from rho-independent transcription terminators of neighboring genes. Although the consensus target site for IS30 is almost palindromic, individual target sites possess considerably less inverted symmetry. In contrast, IS1630 appears to exhibit an increased stringency for inverted repeat recognition, since the majority of target sites had no mismatches in the inverted repeat sequences. In the course of this study, an additional copy of the previously identified insertion sequence ISMi1 was cloned. Analysis of the sequence of this element revealed that the transposase encoded by this element is more than 200 amino acid residues longer and is more closely related to the products of other IS3 family members than had previously been recognized. A potential site for programmed translational frameshifting in ISMi1 was also identified.
Figures






Similar articles
-
Differential posttranslational processing confers intraspecies variation of a major surface lipoprotein and a macrophage-activating lipopeptide of Mycoplasma fermentans.Infect Immun. 1999 Feb;67(2):760-71. doi: 10.1128/IAI.67.2.760-771.1999. Infect Immun. 1999. PMID: 9916088 Free PMC article.
-
IS1550 from Mycoplasma fermentans is transposable in Escherichia coli.FEMS Microbiol Lett. 2001 May 1;198(2):159-64. doi: 10.1111/j.1574-6968.2001.tb10636.x. FEMS Microbiol Lett. 2001. PMID: 11430408
-
Identification of a putative infC-rpmI-rplT operon flanked by long inverted repeats in Mycoplasma fermentans (incognitus strain).Gene. 1993 May 15;127(1):79-85. doi: 10.1016/0378-1119(93)90619-e. Gene. 1993. PMID: 8486291
-
Transposase and cointegrase: specialized transposition proteins of the bacterial insertion sequence IS21 and related elements.Cell Mol Life Sci. 2001 Mar;58(3):403-19. doi: 10.1007/PL00000866. Cell Mol Life Sci. 2001. PMID: 11315188 Free PMC article. Review.
-
Insertion sequences.Microbiol Mol Biol Rev. 1998 Sep;62(3):725-74. doi: 10.1128/MMBR.62.3.725-774.1998. Microbiol Mol Biol Rev. 1998. PMID: 9729608 Free PMC article. Review.
Cited by
-
Molecular genetic analysis of ICEF, an integrative conjugal element that is present as a repetitive sequence in the chromosome of Mycoplasma fermentans PG18.J Bacteriol. 2002 Dec;184(24):6929-41. doi: 10.1128/JB.184.24.6929-6941.2002. J Bacteriol. 2002. PMID: 12446643 Free PMC article.
-
ISAfe1, an ISL3 family insertion sequence from Acidithiobacillus ferrooxidans ATCC 19859.J Bacteriol. 2001 Jul;183(14):4323-9. doi: 10.1128/JB.183.14.4323-4329.2001. J Bacteriol. 2001. PMID: 11418574 Free PMC article.
-
Structural organization and functional properties of miniature DNA insertion sequences in yersiniae.J Bacteriol. 2006 Nov;188(22):7876-84. doi: 10.1128/JB.00942-06. Epub 2006 Sep 8. J Bacteriol. 2006. PMID: 16963573 Free PMC article.
-
Plasticity of the P junc promoter of ISEc11, a new insertion sequence of the IS1111 family.J Bacteriol. 2006 Jul;188(13):4681-9. doi: 10.1128/JB.00332-06. J Bacteriol. 2006. PMID: 16788177 Free PMC article.
-
Genome sequence of the repetitive-sequence-rich Mycoplasma fermentans strain M64.J Bacteriol. 2011 Aug;193(16):4302-3. doi: 10.1128/JB.05228-11. Epub 2011 Jun 3. J Bacteriol. 2011. PMID: 21642450 Free PMC article.
References
-
- Bhugra B, Dybvig K. Identification and characterization of IS1138, a transposable element from Mycoplasma pulmonis that belongs to the IS3 family. Mol Microbiol. 1993;7:577–584. - PubMed
-
- Calcutt, M. J., M. S. Lewis, and K. S. Wise. 1999. Unpublished data.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

