Mutations in the alpha-tubulin 67C gene specifically impair achiasmate segregation in Drosophila melanogaster
- PMID: 10601329
- PMCID: PMC2168102
- DOI: 10.1083/jcb.147.6.1137
Mutations in the alpha-tubulin 67C gene specifically impair achiasmate segregation in Drosophila melanogaster
Abstract
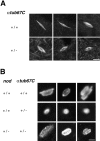
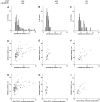
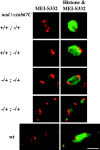
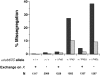
Drosophila melanogaster oocytes heterozygous for mutations in the alpha-tubulin 67C gene (alphatub67C) display defects in centromere positioning during prometaphase of meiosis I. The centromeres do not migrate to the poleward edges of the chromatin mass, and the chromatin fails to stretch during spindle lengthening. These results suggest that the poleward forces acting at the kinetochore are compromised in the alphatub67C mutants. Genetic studies demonstrate that these mutations also strongly and specifically decrease the fidelity of achiasmate chromosome segregation. Proper centromere orientation, chromatin elongation, and faithful segregation can all be restored by a decrease in the amount of the Nod chromokinesin. These results suggest that the accurate segregation of achiasmate chromosomes requires the proper balancing of forces acting on the chromosomes during prometaphase.
Figures
Similar articles
-
Female Meiosis: Synapsis, Recombination, and Segregation in Drosophila melanogaster.Genetics. 2018 Mar;208(3):875-908. doi: 10.1534/genetics.117.300081. Genetics. 2018. PMID: 29487146 Free PMC article. Review.
-
Gamma-tubulin is required for bipolar spindle assembly and for proper kinetochore microtubule attachments during prometaphase I in Drosophila oocytes.PLoS Genet. 2011 Aug;7(8):e1002209. doi: 10.1371/journal.pgen.1002209. Epub 2011 Aug 11. PLoS Genet. 2011. PMID: 21852952 Free PMC article.
-
Genetic variation in rates of nondisjunction: association of two naturally occurring polymorphisms in the chromokinesin nod with increased rates of nondisjunction in Drosophila melanogaster.Genetics. 1999 Aug;152(4):1605-14. doi: 10.1093/genetics/152.4.1605. Genetics. 1999. PMID: 10430586 Free PMC article.
-
Requiem for distributive segregation: achiasmate segregation in Drosophila females.Trends Genet. 1993 Sep;9(9):310-7. doi: 10.1016/0168-9525(93)90249-h. Trends Genet. 1993. PMID: 8236460 Review.
-
Meiosis-specific stable binding of augmin to acentrosomal spindle poles promotes biased microtubule assembly in oocytes.PLoS Genet. 2013 Jun;9(6):e1003562. doi: 10.1371/journal.pgen.1003562. Epub 2013 Jun 13. PLoS Genet. 2013. PMID: 23785300 Free PMC article.
Cited by
-
A deficiency screen of the major autosomes identifies a gene (matrimony) that is haplo-insufficient for achiasmate segregation in Drosophila oocytes.Genetics. 2003 Oct;165(2):637-52. doi: 10.1093/genetics/165.2.637. Genetics. 2003. PMID: 14573476 Free PMC article.
-
Heterochromatic threads connect oscillating chromosomes during prometaphase I in Drosophila oocytes.PLoS Genet. 2009 Jan;5(1):e1000348. doi: 10.1371/journal.pgen.1000348. Epub 2009 Jan 23. PLoS Genet. 2009. PMID: 19165317 Free PMC article.
-
Female Meiosis: Synapsis, Recombination, and Segregation in Drosophila melanogaster.Genetics. 2018 Mar;208(3):875-908. doi: 10.1534/genetics.117.300081. Genetics. 2018. PMID: 29487146 Free PMC article. Review.
-
Dynein promotes achiasmate segregation in Schizosaccharomyces pombe.Genetics. 2005 Jun;170(2):581-90. doi: 10.1534/genetics.104.040253. Epub 2005 Mar 31. Genetics. 2005. PMID: 15802518 Free PMC article.
-
Drosophila Nod protein binds preferentially to the plus ends of microtubules and promotes microtubule polymerization in vitro.Mol Biol Cell. 2005 Nov;16(11):5400-9. doi: 10.1091/mbc.e05-06-0582. Epub 2005 Sep 7. Mol Biol Cell. 2005. PMID: 16148044 Free PMC article.
References
-
- Afshar K., Barton N.R., Hawley R.S., Goldstein L.S. DNA binding and meiotic chromosomal localization of the Drosophila nod kinesin-like protein Cell. 81 1995. 129 138a - PubMed
-
- Dernburg A.F., Sedat J.W., Hawley R.S. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell. 1996;86:135–146. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

