Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins
- PMID: 10601346
- PMCID: PMC2168087
- DOI: 10.1083/jcb.147.6.1351
Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins
Abstract
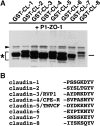
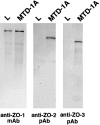
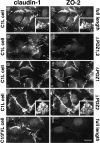
ZO-1, ZO-2, and ZO-3, which contain three PDZ domains (PDZ1 to -3), are concentrated at tight junctions (TJs) in epithelial cells. TJ strands are mainly composed of two distinct types of four-transmembrane proteins, occludin, and claudins, between which occludin was reported to directly bind to ZO-1/ZO-2/ZO-3. However, in occludin-deficient intestinal epithelial cells, ZO-1/ZO-2/ZO-3 were still recruited to TJs. We then examined the possible interactions between ZO-1/ZO-2/ZO-3 and claudins. ZO-1, ZO-2, and ZO-3 bound to the COOH-terminal YV sequence of claudin-1 to -8 through their PDZ1 domains in vitro. Then, claudin-1 or -2 was transfected into L fibroblasts, which express ZO-1 but not ZO-2 or ZO-3. Claudin-1 and -2 were concentrated at cell-cell borders in an elaborate network pattern, to which endogenous ZO-1 was recruited. When ZO-2 or ZO-3 were further transfected, both were recruited to the claudin-based networks together with endogenous ZO-1. Detailed analyses showed that ZO-2 and ZO-3 are recruited to the claudin-based networks through PDZ2 (ZO-2 or ZO-3)/PDZ2 (endogenous ZO-1) and PDZ1 (ZO-2 or ZO-3)/COOH-terminal YV (claudins) interactions. In good agreement, PDZ1 and PDZ2 domains of ZO-1/ZO-2/ZO-3 were also recruited to claudin-based TJs, when introduced into cultured epithelial cells. The possible molecular architecture of TJ plaque structures is discussed.
Figures






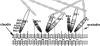
Similar articles
-
Connexin-occludin chimeras containing the ZO-binding domain of occludin localize at MDCK tight junctions and NRK cell contacts.J Cell Biol. 1999 Aug 9;146(3):683-93. doi: 10.1083/jcb.146.3.683. J Cell Biol. 1999. PMID: 10444075 Free PMC article.
-
ZO-3, a novel member of the MAGUK protein family found at the tight junction, interacts with ZO-1 and occludin.J Cell Biol. 1998 Apr 6;141(1):199-208. doi: 10.1083/jcb.141.1.199. J Cell Biol. 1998. PMID: 9531559 Free PMC article.
-
Corneal epithelial tight junctions and their response to lipopolysaccharide challenge.Invest Ophthalmol Vis Sci. 2000 Dec;41(13):4093-100. Invest Ophthalmol Vis Sci. 2000. PMID: 11095601
-
The roles of claudin superfamily proteins in paracellular transport.Traffic. 2001 Feb;2(2):93-8. doi: 10.1034/j.1600-0854.2001.020203.x. Traffic. 2001. PMID: 11247307 Review.
-
Knockout animals and natural mutations as experimental and diagnostic tool for studying tight junction functions in vivo.Biochim Biophys Acta. 2009 Apr;1788(4):813-9. doi: 10.1016/j.bbamem.2008.07.017. Epub 2008 Jul 28. Biochim Biophys Acta. 2009. PMID: 18706387 Review.
Cited by
-
Molecular aspects of intestinal calcium absorption.World J Gastroenterol. 2015 Jun 21;21(23):7142-54. doi: 10.3748/wjg.v21.i23.7142. World J Gastroenterol. 2015. PMID: 26109800 Free PMC article. Review.
-
The N and C termini of ZO-1 are surrounded by distinct proteins and functional protein networks.J Biol Chem. 2013 May 10;288(19):13775-88. doi: 10.1074/jbc.M113.466193. Epub 2013 Apr 3. J Biol Chem. 2013. PMID: 23553632 Free PMC article.
-
Lecture: New light on the role of claudins in the kidney.Organogenesis. 2012 Jan-Mar;8(1):1-9. doi: 10.4161/org.19808. Epub 2012 Jan 1. Organogenesis. 2012. PMID: 22504740 Free PMC article. Review.
-
Nephrin forms a complex with adherens junction proteins and CASK in podocytes and in Madin-Darby canine kidney cells expressing nephrin.Am J Pathol. 2004 Sep;165(3):923-36. doi: 10.1016/S0002-9440(10)63354-8. Am J Pathol. 2004. PMID: 15331416 Free PMC article.
-
Tight junctions in salivary epithelium.J Biomed Biotechnol. 2010;2010:278948. doi: 10.1155/2010/278948. Epub 2010 Feb 18. J Biomed Biotechnol. 2010. PMID: 20182541 Free PMC article. Review.
References
-
- Anderson J.M. Cell signallingMAGUK magic. Curr. Biol. 1996;6:382–384. - PubMed
-
- Balda M.S., Anderson J.M., Matter K. The SH3 domain of the tight junction protein ZO-1 binds to a serine protein kinase that phosphorylates a region C-terminal to this domain. FEBS Lett. 1996;399:326–332. - PubMed
-
- Citi S., Sabanay H., Jakes R., Geiger B., Kendrick-Jones J. Cingulin, a new peripheral component of tight junctions. Nature. 1988;333:272–276. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

