Commitment and differentiation of osteoclast precursor cells by the sequential expression of c-Fms and receptor activator of nuclear factor kappaB (RANK) receptors
- PMID: 10601350
- PMCID: PMC2195707
- DOI: 10.1084/jem.190.12.1741
Commitment and differentiation of osteoclast precursor cells by the sequential expression of c-Fms and receptor activator of nuclear factor kappaB (RANK) receptors
Abstract
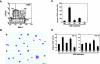
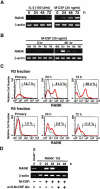
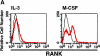
Osteoclasts are terminally differentiated cells derived from hematopoietic stem cells. However, how their precursor cells diverge from macrophagic lineages is not known. We have identified early and late stages of osteoclastogenesis, in which precursor cells sequentially express c-Fms followed by receptor activator of nuclear factor kappaB (RANK), and have demonstrated that RANK expression in early-stage of precursor cells (c-Fms(+)RANK(-)) was stimulated by macrophage colony-stimulating factor (M-CSF). Although M-CSF and RANKL (ligand) induced commitment of late-stage precursor cells (c-Fms(+)RANK(+)) into osteoclasts, even late-stage precursors have the potential to differentiate into macrophages without RANKL. Pretreatment of precursors with M-CSF and delayed addition of RANKL showed that timing of RANK expression and subsequent binding of RANKL are critical for osteoclastogenesis. Thus, the RANK-RANKL system determines the osteoclast differentiation of bipotential precursors in the default pathway of macrophagic differentiation.
Figures
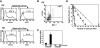







References
-
- Ash P., Loutit J.F., Townsend K.M. Osteoclasts derived from haematopoietic stem cells. Nature. 1980;283:669–670. - PubMed
-
- Scheven B.A., Visser J.W., Nijweide P.J. In vitro osteoclast generation from different bone marrow fractions, including a highly enriched haematopoietic stem cell population. Nature. 1986;321:79–81. - PubMed
-
- Kurihara N., Suda T., Miura Y., Nakauchi H., Kodama H., Hiura K., Hakeda Y., Kumegawa M. Generation of osteoclasts from isolated hematopoietic progenitor cells. Blood. 1989;74:1295–1302. - PubMed
-
- Roodman G.D. Advances in bone biologythe osteoclast. Endocr. Rev. 1996;17:308–332. - PubMed
-
- Manolagas S.C., Jilka R.L. Bone marrow, cytokines, and bone remodeling. Emerging insights into the pathophysiology of osteoporosis. N. Engl. J. Med. 1995;332:305–311. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

