Genetic-biochemical analysis and distribution of the Ambler class A beta-lactamase CME-2, responsible for extended-spectrum cephalosporin resistance in Chryseobacterium (Flavobacterium) meningosepticum
- PMID: 10602714
- PMCID: PMC89619
- DOI: 10.1128/AAC.44.1.1-9.2000
Genetic-biochemical analysis and distribution of the Ambler class A beta-lactamase CME-2, responsible for extended-spectrum cephalosporin resistance in Chryseobacterium (Flavobacterium) meningosepticum
Abstract
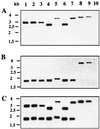
In vitro synergy between extended-spectrum cephalosporins and either clavulanic acid or cefoxitin was found for Chryseobacterium meningosepticum isolates during a double-disk assay on an agar plate. An extended-spectrum beta-lactamase (ESBL) gene from a C. meningosepticum clinical isolate was cloned and expressed in Escherichia coli DH10B. Its protein conferred resistance to most beta-lactams including extended-spectrum cephalosporins but not to cephamycins or to imipenem. Its activity was strongly inhibited by clavulanic acid, sulbactam, and tazobactam, as well as by cephamycins and imipenem. Sequence analysis of the cloned DNA fragment revealed an open reading frame (ORF) of 891 bp with a G+C content of 33.9%, which lies close to the expected range of G+C contents of members of the Chryseobacterium genus. The ORF encoded a precursor protein of 297 amino acids, giving a mature protein with a molecular mass of 31 kDa and a pI value of 9.2 in E. coli. This gene was very likely chromosomally located. Amino acid sequence comparison showed that this beta-lactamase, named CME-2 (C. meningosepticum ESBL), is a novel ESBL of the Ambler class A group (Bush functional group 2be), being weakly related to other class A beta-lactamases. It shares only 39 and 35% identities with the ESBLs VEB-1 from E. coli MG-1 and CBL-A from Bacteroides uniformis, respectively. The distribution of bla(CME-2) among unrelated C. meningosepticum species isolates showed that bla(CME-2)-like genes were found in the C. meningosepticum strains studied but were absent from strains of other C. meningosepticum-related species. Each C. meningosepticum strain produced at least two beta-lactamases, with one of them being a noninducible serine ESBL with variable pIs ranging from 7.0 to 8.5.
Figures



References
-
- Ambler R P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289:321–331. - PubMed
-
- Barthélémy M, Péduzzi J, Bernard H, Tancrède C, Labia R. Close amino-acid sequence relationship between the new plasmid-mediated extended-spectrum beta-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim Biophys Acta. 1992;1122:15–22. - PubMed
-
- Bloch K C, Nadarajah R, Jacobs R. Chryseobacterium meningosepticum: an emerging pathogen among immunocompromised adults. Report of 6 cases and literature review. Medicine (Baltimore) 1997;76:30–41. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

