Better conditions for mammalian in vitro splicing provided by acetate and glutamate as potassium counterions
- PMID: 10606638
- PMCID: PMC102525
- DOI: 10.1093/nar/28.2.416
Better conditions for mammalian in vitro splicing provided by acetate and glutamate as potassium counterions
Abstract
We demonstrate here that replacing potassium chloride (KCl) with potassium acetate (KAc) or potassium glutamate (KGlu) routinely enhances the yield of RNA intermediates and products obtained from in vitro splicing reactions performed in HeLa cell nuclear extract. This effect was reproducibly observed with multiple splicing substrates. The enhanced yields are at least partially due to stabilization of splicing precursors and products in the KAc and KGlu reactions. This stabilization relative to KCl reactions was greatest with KGlu and was observed over an extended potassium concentration range. The RNA stability differences could not be attributed to heavy metal contamination of the KCl, since ultrapure preparations of this salt yielded similar results. After testing various methods for altering the salts, we found that substitution of KAc or KGlu for KCl and MgAc(2)for MgCl(2)in splicing reactions is the simplest and most effective. Since the conditions defined here more closely mimic in vivo ionic concentrations, they may permit the study of more weakly spliced substrates, as well as facilitate more detailed analyses of spliceosome structure and function.
Figures


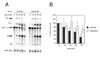
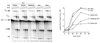

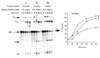
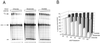
References
-
- Hardy S.F., Grabowski,P.J., Padgett,R.A. and Sharp,P.A. (1984) Nature, 308, 375–377. - PubMed
-
- Krainer A.R., Maniatis,T., Ruskin,B. and Green,M.R. (1984) Cell, 36, 993–1005. - PubMed
-
- Hernandez N. and Keller,W. (1983) Cell, 35, 89–99. - PubMed
-
- Kramer A. and Keller,W. (1990) In Dahlberg,J.E. and Abelson,J.N. (eds), Methods in Enzymology— RNA Processing, Part B: Specific Methods, Vol. 181. Academic Press, San Diego, CA, pp. 3–11.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

