Quasispecies development of Helicobacter pylori observed in paired isolates obtained years apart from the same host
- PMID: 10608776
- PMCID: PMC2766531
- DOI: 10.1086/315173
Quasispecies development of Helicobacter pylori observed in paired isolates obtained years apart from the same host
Abstract
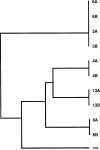
Helicobacter pylori isolates show greater genetic diversity than other bacterial species studied, but the basis for this phenomenon is unknown. Whether detectable genomic mutation appears within an H. pylori population during persistent colonization was investigated. Paired H. pylori populations obtained across 7- to 10-year intervals from 13 patients were characterized by use of methods including polymerase chain reaction (PCR) genotyping for cagA, vacA, iceA, recA, and IS605; random arbitrarily primed DNA (RAPD)-PCR and amplified fragment length polymorphism (AFLP) analysis; and ELISA, to determine Lewis phenotypes. Genotyping, including recA sequence analysis, revealed that initial and follow-up populations represented the same population in 11 patients (85%). Nevertheless, distinct dissimilarities were shown within each of these 11 pairs by both RAPD-PCR and AFLP analyses. During follow-up, Lewis-y levels, but not Lewis-x levels, decreased significantly. The changes detected by RAPD-PCR and AFLP indicate that genetic drift occurs within H. pylori populations over the course of years of colonization of a single host.
Figures



References
-
- Logan RPH, Berg DE. Genetic diversity of Helicobacter pylori. Lancet. 1996;348:1462–3. - PubMed
-
- Kansau I, Raymond J, Bingen E, et al. Genotyping of Helicobacter pylori isolates by sequencing of PCR products and comparison with the RAPD technique. Res Microbiol. 1996;147:661–9. - PubMed
-
- Akopyants NS, Clifton SW, Kersulyte D, et al. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol. 1998;28:37–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

