Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters
- PMID: 10611242
- PMCID: PMC85153
- DOI: 10.1128/MCB.20.2.634-647.2000
Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters
Abstract
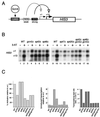
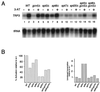
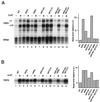
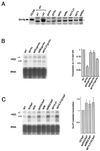
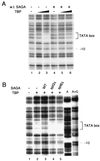
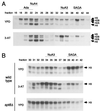
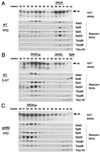
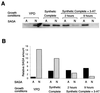
SAGA is a 1.8-MDa yeast protein complex that is composed of several distinct classes of transcription-related factors, including the adaptor/acetyltransferase Gcn5, Spt proteins, and a subset of TBP-associated factors. Our results indicate that mutations that completely disrupt SAGA (deletions of SPT7 or SPT20) strongly reduce transcriptional activation at the HIS3 and TRP3 genes and that Gcn5 is required for normal HIS3 transcriptional start site selection. Surprisingly, mutations in Spt proteins involved in the SAGA-TBP interaction (Spt3 and Spt8) cause derepression of HIS3 and TRP3 transcription in the uninduced state. Consistent with this finding, wild-type SAGA inhibits TBP binding to the HIS3 promoter in vitro, while SAGA lacking Spt3 or Spt8 is not inhibitory. We detected two distinct forms of SAGA in cell extracts and, strikingly, one lacks Spt8. Conditions that induce HIS3 and TRP3 transcription result in an altered balance between these complexes strongly in favor of the form without Spt8. These results suggest that the composition of SAGA may be dynamic in vivo and may be regulated through dissociable inhibitory subunits.
Figures


References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994.
-
- Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
-
- Barlev N A, Candau R, Wang L, Darpino P, Silverman N, Berger S L. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. - PubMed
-
- Berger S L, Piña B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992;70:251–265. - PubMed
-
- Birck C, Poch O, Romier C, Ruff M, Mengus G, Lavigne A-C, Davidson I, Moras D. Human TAFII28 and TAFII18 interact through a canonical histone fold encoded by atypical evolutionary conserved sequence motifs also found in the SPT3 TAFII family. Cell. 1998;94:239–249. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
