Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA
- PMID: 10611295
- PMCID: PMC24730
- DOI: 10.1073/pnas.96.26.14813
Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA
Abstract
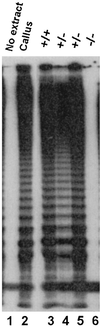
Telomerase is an essential enzyme that maintains telomeres on eukaryotic chromosomes. In mammals, telomerase is required for the lifelong proliferative capacity of normal regenerative and reproductive tissues and for sustained growth in a dedifferentiated state. Although the importance of telomeres was first elucidated in plants 60 years ago, little is known about the role of telomeres and telomerase in plant growth and development. Here we report the cloning and characterization of the Arabidopsis telomerase reverse transcriptase (TERT) gene, AtTERT. AtTERT is predicted to encode a highly basic protein of 131 kDa that harbors the reverse transcriptase and telomerase-specific motifs common to all known TERT proteins. AtTERT mRNA is 10-20 times more abundant in callus, which has high levels of telomerase activity, versus leaves, which contain no detectable telomerase. Plants homozygous for a transfer DNA insertion into the AtTERT gene lack telomerase activity, confirming the identity and function of this gene. Because telomeres in wild-type Arabidopsis are short, the discovery that telomerase-null plants are viable for at least two generations was unexpected. In the absence of telomerase, telomeres decline by approximately 500 bp per generation, a rate 10 times slower than seen in telomerase-deficient mice. This gradual loss of telomeric DNA may reflect a reduced rate of nucleotide depletion per round of DNA replication, or the requirement for fewer cell divisions per organismal generation. Nevertheless, progressive telomere shortening in the mutants, however slow, ultimately should be lethal.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

