Pressure-induced protein-folding/unfolding kinetics
- PMID: 10611301
- PMCID: PMC24736
- DOI: 10.1073/pnas.96.26.14848
Pressure-induced protein-folding/unfolding kinetics
Abstract
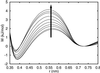
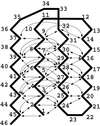
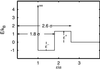
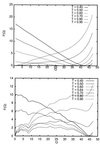
We use an off-lattice minimalist model to describe the effects of pressure in slowing down the folding/unfolding kinetics of proteins when subjected to increasingly larger pressures. The potential energy function used to describe the interactions between beads in the model includes the effects of pressure on the pairwise interaction of hydrophobic groups in water. We show that pressure affects the participation of contacts in the transition state. More significantly, pressure exponentially decreases the chain reconfigurational diffusion coefficient. These results are consistent with experimental results on the kinetics of pressure-denaturation of staphylococcal nuclease.
Figures



Similar articles
-
High-pressure denaturation of staphylococcal nuclease proline-to-glycine substitution mutants.Biochemistry. 1996 Mar 26;35(12):3857-64. doi: 10.1021/bi952012g. Biochemistry. 1996. PMID: 8620010
-
Structural characterization of the pressure-denatured state and unfolding/refolding kinetics of staphylococcal nuclease by synchrotron small-angle X-ray scattering and Fourier-transform infrared spectroscopy.J Mol Biol. 1998 Jan 16;275(2):389-402. doi: 10.1006/jmbi.1997.1454. J Mol Biol. 1998. PMID: 9466917
-
Studying pressure denaturation of a protein by molecular dynamics simulations.Proteins. 2010 May 15;78(7):1641-51. doi: 10.1002/prot.22680. Proteins. 2010. PMID: 20146357
-
Thermodynamics of denaturation of staphylococcal nuclease mutants: an intermediate state in protein folding.FASEB J. 1996 Jan;10(1):67-74. doi: 10.1096/fasebj.10.1.8566550. FASEB J. 1996. PMID: 8566550 Review.
-
Staphylococcal nuclease: a showcase of m-value effects.Adv Protein Chem. 1995;46:217-47. doi: 10.1016/s0065-3233(08)60336-8. Adv Protein Chem. 1995. PMID: 7771319 Review. No abstract available.
Cited by
-
Critical phenomena in the temperature-pressure-crowding phase diagram of a protein.Phys Rev X. 2019 Oct-Dec;9(4):041035. doi: 10.1103/physrevx.9.041035. Epub 2019 Nov 18. Phys Rev X. 2019. PMID: 32642303 Free PMC article.
-
Solvation in protein folding analysis: combination of theoretical and experimental approaches.Proc Natl Acad Sci U S A. 2004 Mar 2;101(9):2834-9. doi: 10.1073/pnas.0304180101. Epub 2004 Feb 20. Proc Natl Acad Sci U S A. 2004. PMID: 14978284 Free PMC article.
-
Chevron behavior and isostable enthalpic barriers in protein folding: successes and limitations of simple Gō-like modeling.Biophys J. 2005 Jul;89(1):520-35. doi: 10.1529/biophysj.104.057471. Epub 2005 Apr 29. Biophys J. 2005. PMID: 15863486 Free PMC article.
-
Protein folding mediated by solvation: water expulsion and formation of the hydrophobic core occur after the structural collapse.Proc Natl Acad Sci U S A. 2002 Jan 22;99(2):685-90. doi: 10.1073/pnas.022387699. Proc Natl Acad Sci U S A. 2002. PMID: 11805324 Free PMC article.
-
High-Pressure-Driven Reversible Dissociation of α-Synuclein Fibrils Reveals Structural Hierarchy.Biophys J. 2017 Oct 17;113(8):1685-1696. doi: 10.1016/j.bpj.2017.08.042. Biophys J. 2017. PMID: 29045863 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

